The origin of bladder cancer from mucosal field effects
- PMID: 35747385
- PMCID: PMC9209726
- DOI: 10.1016/j.isci.2022.104551
The origin of bladder cancer from mucosal field effects
Erratum in
-
Erratum: The origin of bladder cancer from mucosal field effects.iScience. 2022 Jul 4;25(7):104715. doi: 10.1016/j.isci.2022.104715. eCollection 2022 Jul 15. iScience. 2022. PMID: 35811851 Free PMC article.
Abstract
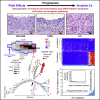
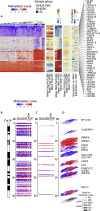
Whole-organ mapping was used to study molecular changes in the evolution of bladder cancer from field effects. We identified more than 100 dysregulated pathways, involving immunity, differentiation, and transformation, as initiators of carcinogenesis. Dysregulation of interleukins signified the involvement of inflammation in the incipient phases of the process. An aberrant methylation/expression of multiple HOX genes signified dysregulation of the differentiation program. We identified three types of mutations based on their geographic distribution. The most common were mutations restricted to individual mucosal samples that targeted uroprogenitor cells. Two types of mutations were associated with clonal expansion and involved large areas of mucosa. The α mutations occurred at low frequencies while the β mutations increased in frequency with disease progression. Modeling revealed that bladder carcinogenesis spans 10-15 years and can be divided into dormant and progressive phases. The progressive phase lasted 1-2 years and was driven by β mutations.
Keywords: Cancer; Cell biology; Molecular biology.
© 2022 The Author(s).
Conflict of interest statement
We confirm that there are no competing interests.
Figures







References
-
- Aryee M.J., Jaffe A.E., Corrada-Bravo H., Ladd-Acosta C., Feinberg A.P., Hansen K.D., Irizarry R.A. Minfi: a flexible and comprehensive Bioconductor package for the analysis of Infinium DNA methylation microarrays. Bioinformatics. 2014;30:1363–1369. doi: 10.1093/bioinformatics/btu049. - DOI - PMC - PubMed
-
- Benjamini Y., Hochberg Y. Controlling the false discovery rate - a practical and powerful approach to multiple testing. J. Roy. Stat. Soc. B Stat. Methodol. 1995;57:289–300. doi: 10.1111/j.2517-6161.1995.tb02031.x. - DOI
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

