Transcriptional Regulation of ING5 and its Suppressive Effects on Gastric Cancer
- PMID: 35747809
- PMCID: PMC9209732
- DOI: 10.3389/fonc.2022.918954
Transcriptional Regulation of ING5 and its Suppressive Effects on Gastric Cancer
Abstract
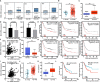
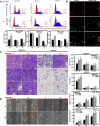
ING5 targets histone acetyltransferase or histone deacetylase complexes for local chromatin remodeling. Its transcriptional regulation and suppressive effects on gastric cancer remain elusive. Luciferase assay, EMSA, and ChIP were used to identify the cis-acting elements and trans-acting factors of the ING5 gene. We analyzed the effects of SAHA on the aggressive phenotypes of ING5 transfectants, and the effects of different ING5 mutants on aggressive phenotypes in SGC-7901 cells. Finally, we observed the effects of ING5 abrogation on gastric carcinogenesis. EMSA and ChIP showed that both SRF (-717 to -678 bp) and YY1 (-48 to 25bp) interacted with the promoter of ING5 and up-regulated ING5 expression in gastric cancer via SRF-YY1-ING5-p53 complex formation. ING5, SRF, and YY1 were overexpressed in gastric cancer, (P<0.05), and associated with worse prognosis of gastric cancer patients (P<0.05). ING5 had positive relationships with SRF and YY1 expression in gastric cancer (P<0.05). SAHA treatment caused early arrest at S phase in ING5 transfectants of SGC-7901 (P<0.05), and either 0.5 or 1.0 μM SAHA enhanced their migration and invasion (P<0.05). The wild-type and mutant ING5 transfectants showed lower viability and invasion than the control (P<0.05) with low CDC25, VEGF, and MMP-9 expression. Gastric spontaneous adenocarcinoma was observed in Atp4b-cre; ING5f/f, Pdx1-cre; ING5f/f, and K19-cre; ING5f/f mice. ING5 deletion increased the sensitivity of MNU-induced gastric carcinogenesis. ING5 mRNA might be a good marker of gastric carcinogenesis, and poor prognosis. ING5 expression was positively regulated by the interaction of SRF-YY1-ING5-p53 complex within the ING5 promoter from -50 bp upstream to the transcription start site. ING5 deletion might contribute to the tumorigenesis and histogenesis of gastric cancer.
Keywords: ING5; gastric cancer; transcriptional regulation; tumor suppressor; tumorigenesis.
Copyright © 2022 Zheng, Xue, Wu, Xu, Zhao and Cui.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures




Similar articles
-
The roles of ING5 in cancer: A tumor suppressor.Front Cell Dev Biol. 2022 Nov 8;10:1012179. doi: 10.3389/fcell.2022.1012179. eCollection 2022. Front Cell Dev Biol. 2022. PMID: 36425530 Free PMC article. Review.
-
ING5 suppresses proliferation, apoptosis, migration and invasion, and induces autophagy and differentiation of gastric cancer cells: a good marker for carcinogenesis and subsequent progression.Oncotarget. 2015 Aug 14;6(23):19552-79. doi: 10.18632/oncotarget.3735. Oncotarget. 2015. PMID: 25980581 Free PMC article.
-
The roles of ING5 in gliomas: a good marker for tumorigenesis and a potential target for gene therapy.Oncotarget. 2017 May 11;8(34):56558-56568. doi: 10.18632/oncotarget.17802. eCollection 2017 Aug 22. Oncotarget. 2017. PMID: 28915612 Free PMC article.
-
The altered expression of ING5 protein is involved in gastric carcinogenesis and subsequent progression.Hum Pathol. 2011 Jan;42(1):25-35. doi: 10.1016/j.humpath.2010.05.024. Epub 2010 Nov 9. Hum Pathol. 2011. PMID: 21062663
-
The down-regulated ING5 expression in lung cancer: a potential target of gene therapy.Oncotarget. 2016 Aug 23;7(34):54596-54615. doi: 10.18632/oncotarget.10519. Oncotarget. 2016. PMID: 27409347 Free PMC article.
Cited by
-
Bioinformatics Analysis of the Expression and Prognostic Significance of Transcription Factor YY1 in Gastric Cancer.Cancer Rep (Hoboken). 2025 Mar;8(3):e70181. doi: 10.1002/cnr2.70181. Cancer Rep (Hoboken). 2025. PMID: 40088083 Free PMC article.
-
The Antitumor and Sorafenib-resistant Reversal Effects of Ursolic Acid on Hepatocellular Carcinoma via Targeting ING5.Int J Biol Sci. 2024 Aug 5;20(11):4190-4208. doi: 10.7150/ijbs.97720. eCollection 2024. Int J Biol Sci. 2024. PMID: 39247819 Free PMC article.
-
The roles of ING5 in cancer: A tumor suppressor.Front Cell Dev Biol. 2022 Nov 8;10:1012179. doi: 10.3389/fcell.2022.1012179. eCollection 2022. Front Cell Dev Biol. 2022. PMID: 36425530 Free PMC article. Review.
References
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

