DNASE1L3 inhibits proliferation, invasion and metastasis of hepatocellular carcinoma by interacting with β-catenin to promote its ubiquitin degradation pathway
- PMID: 35748106
- PMCID: PMC9436914
- DOI: 10.1111/cpr.13273
DNASE1L3 inhibits proliferation, invasion and metastasis of hepatocellular carcinoma by interacting with β-catenin to promote its ubiquitin degradation pathway
Abstract
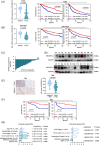
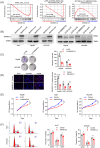
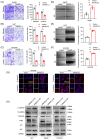
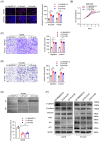
As a member of the deoxyribonuclease 1 family, DNASE1L3 plays a significant role both inside and outside the cell. However, the role of DNASE1L3 in hepatocellular carcinoma (HCC) and its molecular basis remains to be further investigated. In this study, we report that DNASE1L3 is downregulated in clinical HCC samples and evaluate the relationship between its expression and HCC clinical features. In vivo and in vitro experiments showed that DNASE1L3 negatively regulates the proliferation, invasion and metastasis of HCC cells. Mechanistic studies showed that DNASE1L3 recruits components of the cytoplasmic β-catenin destruction complex (GSK-3β and Axin), promotes the ubiquitination degradation of β-catenin, and inhibits its nuclear transfer, thus, decreasing c-Myc, P21 and P27 level. Ultimately, cell cycle and EMT signals are restrained. In general, this study provides new insight into the mechanism for HCC and suggests that DNASE1L3 can become a considerable target for HCC.
© 2022 The Authors. Cell Proliferation published by European Cell Proliferation Society and John Wiley & Sons Ltd.
Conflict of interest statement
The authors declare no conflict of interests.
Figures




Similar articles
-
Spindle and kinetochore-associated protein 2 facilitates the proliferation and invasion of hepatocellular carcinoma via the regulation of Wnt/β-catenin signaling.Exp Cell Res. 2020 Oct 1;395(1):112181. doi: 10.1016/j.yexcr.2020.112181. Epub 2020 Jul 15. Exp Cell Res. 2020. PMID: 32682011
-
ARHGAP17 Inhibits Hepatocellular Carcinoma Progression by Inactivation of Wnt/β-Catenin Signaling Pathway.Biochem Genet. 2025 Jun;63(3):2299-2311. doi: 10.1007/s10528-024-10822-5. Epub 2024 May 9. Biochem Genet. 2025. PMID: 38724713
-
Silencing of lemur tyrosine kinase 2 restricts the proliferation and invasion of hepatocellular carcinoma through modulation of GSK-3β/Wnt/β-catenin signaling.Biochem Biophys Res Commun. 2019 Oct 1;517(4):722-728. doi: 10.1016/j.bbrc.2019.07.122. Epub 2019 Aug 5. Biochem Biophys Res Commun. 2019. PMID: 31395338
-
Pivotal roles of glycogen synthase-3 in hepatocellular carcinoma.Adv Biol Regul. 2017 Aug;65:59-76. doi: 10.1016/j.jbior.2017.06.002. Epub 2017 Jun 6. Adv Biol Regul. 2017. PMID: 28619606 Review.
-
Dysregulation of Wnt/β-catenin signaling by protein kinases in hepatocellular carcinoma and its therapeutic application.Cancer Sci. 2021 May;112(5):1695-1706. doi: 10.1111/cas.14861. Epub 2021 Apr 6. Cancer Sci. 2021. PMID: 33605517 Free PMC article. Review.
Cited by
-
The regulatory role of the circELMOD3-associated ceRNA network in the progression and prognosis of hepatocellular carcinoma.Front Genet. 2025 Apr 15;16:1521360. doi: 10.3389/fgene.2025.1521360. eCollection 2025. Front Genet. 2025. PMID: 40303979 Free PMC article.
-
Pan‑cancer analysis of the deoxyribonuclease gene family.Mol Clin Oncol. 2023 Feb 2;18(3):19. doi: 10.3892/mco.2023.2615. eCollection 2023 Mar. Mol Clin Oncol. 2023. PMID: 36798465 Free PMC article.
-
A novel prognostic signature based on immunogenic cell death score predicts outcomes and response to transcatheter arterial chemoembolization and immunotherapy in hepatocellular carcinoma.J Cancer Res Clin Oncol. 2023 Oct;149(13):11411-11429. doi: 10.1007/s00432-023-05017-1. Epub 2023 Jun 29. J Cancer Res Clin Oncol. 2023. PMID: 37382674 Free PMC article.
-
Constructing a prognostic model for hepatocellular carcinoma based on bioinformatics analysis of inflammation-related genes.Front Med (Lausanne). 2024 Jul 11;11:1420353. doi: 10.3389/fmed.2024.1420353. eCollection 2024. Front Med (Lausanne). 2024. PMID: 39055701 Free PMC article.
-
Identifying pathogenic variants related to systemic lupus erythematosus by integrating genomic databases and a bioinformatic approach.Genomics Inform. 2023 Sep;21(3):e37. doi: 10.5808/gi.23002. Epub 2023 Sep 27. Genomics Inform. 2023. PMID: 37813633 Free PMC article.
References
-
- Global Burden of Disease Liver Cancer Collaboration , Akinyemiju T, Abera S, et al. The burden of primary liver cancer and underlying etiologies from 1990 to 2015 at the global, regional, and National Level: results from the global burden of disease study 2015. JAMA Oncol. 2017;3(12):1683‐1691. doi:10.1001/jamaoncol.2017.3055 - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials

