TMED3 Complex Mediates ER Stress-Associated Secretion of CFTR, Pendrin, and SARS-CoV-2 Spike
- PMID: 35748162
- PMCID: PMC9350134
- DOI: 10.1002/advs.202105320
TMED3 Complex Mediates ER Stress-Associated Secretion of CFTR, Pendrin, and SARS-CoV-2 Spike
Abstract
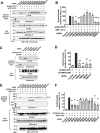
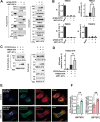
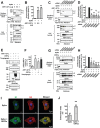
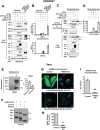
Under ER stress conditions, the ER form of transmembrane proteins can reach the plasma membrane via a Golgi-independent unconventional protein secretion (UPS) pathway. However, the targeting mechanisms of membrane proteins for UPS are unknown. Here, this study reports that TMED proteins play a critical role in the ER stress-associated UPS of transmembrane proteins. The gene silencing results reveal that TMED2, TMED3, TMED9 and TMED10 are involved in the UPS of transmembrane proteins, such as CFTR, pendrin and SARS-CoV-2 Spike. Subsequent mechanistic analyses indicate that TMED3 recognizes the ER core-glycosylated protein cargos and that the heteromeric TMED2/3/9/10 complex mediates their UPS. Co-expression of all four TMEDs improves, while each single expression reduces, the UPS and ion transport function of trafficking-deficient ΔF508-CFTR and p.H723R-pendrin, which cause cystic fibrosis and Pendred syndrome, respectively. In contrast, TMED2/3/9/10 silencing reduces SARS-CoV-2 viral release. These results provide evidence for a common role of TMED3 and related TMEDs in the ER stress-associated, Golgi-independent secretion of transmembrane proteins.
Keywords: CFTR; SARS-CoV-2 spike; TMED; UPS; pendrin.
© 2022 The Authors. Advanced Science published by Wiley-VCH GmbH.
Conflict of interest statement
The authors declare no conflict of interest.
Figures





References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous
