m6A in the Signal Transduction Network
- PMID: 35748227
- PMCID: PMC9260138
- DOI: 10.14348/molcells.2022.0017
m6A in the Signal Transduction Network
Abstract
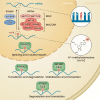
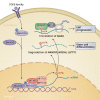
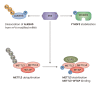
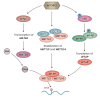
In response to environmental changes, signaling pathways rewire gene expression programs through transcription factors. Epigenetic modification of the transcribed RNA can be another layer of gene expression regulation. N6-adenosine methylation (m6A) is one of the most common modifications on mRNA. It is a reversible chemical mark catalyzed by the enzymes that deposit and remove methyl groups. m6A recruits effector proteins that determine the fate of mRNAs through changes in splicing, cellular localization, stability, and translation efficiency. Emerging evidence shows that key signal transduction pathways including TGFβ (transforming growth factor-β), ERK (extracellular signal-regulated kinase), and mTORC1 (mechanistic target of rapamycin complex 1) regulate downstream gene expression through m6A processing. Conversely, m6A can modulate the activity of signal transduction networks via m6A modification of signaling pathway genes or by acting as a ligand for receptors. In this review, we discuss the current understanding of the crosstalk between m6A and signaling pathways and its implication for biological systems.
Keywords:
ERK;
Conflict of interest statement
The authors have no potential conflicts of interest to disclose.
Figures
Similar articles
-
mTORC1 stimulates cell growth through SAM synthesis and m6A mRNA-dependent control of protein synthesis.Mol Cell. 2021 May 20;81(10):2076-2093.e9. doi: 10.1016/j.molcel.2021.03.009. Epub 2021 Mar 22. Mol Cell. 2021. PMID: 33756106 Free PMC article.
-
The crosstalk between RNA m6A epitranscriptome and TGFβ signaling pathway contributes to the arrest of cell cycle.Gene. 2020 May 15;738:144483. doi: 10.1016/j.gene.2020.144483. Epub 2020 Feb 15. Gene. 2020. PMID: 32070750
-
mTORC1 promotes cell growth via m6A-dependent mRNA degradation.Mol Cell. 2021 May 20;81(10):2064-2075.e8. doi: 10.1016/j.molcel.2021.03.010. Epub 2021 Mar 22. Mol Cell. 2021. PMID: 33756105 Free PMC article.
-
N6-methyladenosine modification in mRNA: machinery, function and implications for health and diseases.FEBS J. 2016 May;283(9):1607-30. doi: 10.1111/febs.13614. Epub 2015 Dec 31. FEBS J. 2016. PMID: 26645578 Review.
-
Epitranscriptomics in the Heart: a Focus on m6A.Curr Heart Fail Rep. 2020 Oct;17(5):205-212. doi: 10.1007/s11897-020-00473-z. Curr Heart Fail Rep. 2020. PMID: 32813261 Free PMC article. Review.
Cited by
-
The Epitranscriptomic Mechanism of Metal Toxicity and Carcinogenesis.Int J Mol Sci. 2022 Oct 5;23(19):11830. doi: 10.3390/ijms231911830. Int J Mol Sci. 2022. PMID: 36233132 Free PMC article. Review.
-
RBM15‑mediating MDR1 mRNA m6A methylation regulated by the TGF‑β signaling pathway in paclitaxel‑resistant ovarian cancer.Int J Oncol. 2023 Oct;63(4):112. doi: 10.3892/ijo.2023.5560. Epub 2023 Aug 18. Int J Oncol. 2023. PMID: 37594126 Free PMC article.
-
Regulation of m6A Methylome in Cancer: Mechanisms, Implications, and Therapeutic Strategies.Cells. 2023 Dec 28;13(1):66. doi: 10.3390/cells13010066. Cells. 2023. PMID: 38201270 Free PMC article. Review.
-
Unraveling the genetic and epigenetic landscape governing intramuscular fat deposition in rabbits: Insights and implications.Food Chem (Oxf). 2024 Aug 30;9:100222. doi: 10.1016/j.fochms.2024.100222. eCollection 2024 Dec 30. Food Chem (Oxf). 2024. PMID: 39290671 Free PMC article. Review.
-
Crosstalk among N6-methyladenosine modification and RNAs in central nervous system injuries.Front Cell Neurosci. 2022 Sep 29;16:1013450. doi: 10.3389/fncel.2022.1013450. eCollection 2022. Front Cell Neurosci. 2022. PMID: 36246528 Free PMC article. Review.
References
-
- Bokar J.A., Rath-Shambaugh M.E., Ludwiczak R., Narayan P., Rottman F. Characterization and partial purification of mRNA N6-adenosine methyltransferase from HeLa cell nuclei. Internal mRNA methylation requires a multisubunit complex. J. Biol. Chem. 1994;269:17697–17704. doi: 10.1016/S0021-9258(17)32497-3. - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

