Deciphering signal transduction networks in the liver by mechanistic mathematical modelling
- PMID: 35748700
- PMCID: PMC9246346
- DOI: 10.1042/BCJ20210548
Deciphering signal transduction networks in the liver by mechanistic mathematical modelling
Abstract
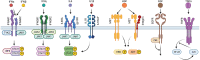
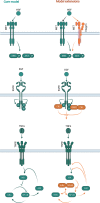
In health and disease, liver cells are continuously exposed to cytokines and growth factors. While individual signal transduction pathways induced by these factors were studied in great detail, the cellular responses induced by repeated or combined stimulations are complex and less understood. Growth factor receptors on the cell surface of hepatocytes were shown to be regulated by receptor interactions, receptor trafficking and feedback regulation. Here, we exemplify how mechanistic mathematical modelling based on quantitative data can be employed to disentangle these interactions at the molecular level. Crucial is the analysis at a mechanistic level based on quantitative longitudinal data within a mathematical framework. In such multi-layered information, step-wise mathematical modelling using submodules is of advantage, which is fostered by sharing of standardized experimental data and mathematical models. Integration of signal transduction with metabolic regulation in the liver and mechanistic links to translational approaches promise to provide predictive tools for biology and personalized medicine.
Keywords: cytokines; growth factors; hepatocellular carcinoma; hepatocytes; liver; mathematical modelling.
© 2022 The Author(s).
Conflict of interest statement
The authors declare that there are no competing interests associated with the manuscript.
Figures

References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources

