Cd59 and inflammation regulate Schwann cell development
- PMID: 35748863
- PMCID: PMC9232220
- DOI: 10.7554/eLife.76640
Cd59 and inflammation regulate Schwann cell development
Abstract
Efficient neurotransmission is essential for organism survival and is enhanced by myelination. However, the genes that regulate myelin and myelinating glial cell development have not been fully characterized. Data from our lab and others demonstrates that cd59, which encodes for a small GPI-anchored glycoprotein, is highly expressed in developing zebrafish, rodent, and human oligodendrocytes (OLs) and Schwann cells (SCs), and that patients with CD59 dysfunction develop neurological dysfunction during early childhood. Yet, the function of Cd59 in the developing nervous system is currently undefined. In this study, we demonstrate that cd59 is expressed in a subset of developing SCs. Using cd59 mutant zebrafish, we show that developing SCs proliferate excessively and nerves may have reduced myelin volume, altered myelin ultrastructure, and perturbed node of Ranvier assembly. Finally, we demonstrate that complement activity is elevated in cd59 mutants and that inhibiting inflammation restores SC proliferation, myelin volume, and nodes of Ranvier to wildtype levels. Together, this work identifies Cd59 and developmental inflammation as key players in myelinating glial cell development, highlighting the collaboration between glia and the innate immune system to ensure normal neural development.
Keywords: CD59; Schwann cell; complement; developmental biology; inflammation; myelin; neuroscience; node of Ranvier; zebrafish.
Plain language summary
The nervous system of vertebrates is made of up of complex networks of nerve cells that send signals to one another. In addition to these cells, there are a number of supporting cells that help nerves carry out their role. Schwann cells, for example, help nerve cells to transmit information faster by wrapping their long extensions in a fatty membrane called myelin. When myelin is not produced properly, this can disturb the signals between nerve cells, leading to neurological defects. Schwann cells mature simultaneously with nerve cells in the embryo. However, it was not fully understood how Schwann cells generate myelin during development. To investigate, Wiltbank et al. studied the embryos of zebrafish, which, unlike other vertebrates, are transparent and develop outside of the womb. These qualities make it easier to observe how cells in the nervous system grow in real-time using a microscope. First, the team analyzed genetic data collected from the embryo of zebrafish and mice to search for genes that are highly abundant in Schwann cells during development. This led to the discovery of a gene called cd59, which codes for a protein that also interacts with the immune system. To find out whether Schwann cells rely on cd59, Wiltbank et al. deleted the cd59 gene in zebrafish embryos. Without cd59, the Schwann cells produced too many copies of themselves and this, in turn, impaired the appropriate production of myelin. Since the protein encoded by cd59 normally balances inflammation levels, it was possible that losing this gene overactivated the immune system in the zebrafish embryos. In support of this hypothesis, when the cd59 mutant embryos were treated with an anti-inflammatory drug, this corrected Schwann cell overproduction and restored myelin formation. Taken together, these findings reveal how inflammation helps determine the number of Schwann cells produced during development, and that cd59 prevents this process from getting carried away. This suggests that the nervous system and immune system may work together to build the nervous system. In the future, it will be interesting to investigate whether cd59 acts in a similar way during the development of the human nervous system.
© 2022, Wiltbank et al.
Conflict of interest statement
AW, ES, SC, MP, SK No competing interests declared
Figures



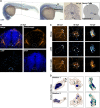

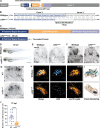

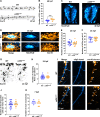







References
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

