Convergent evolution of antiviral machinery derived from endogenous retrovirus truncated envelope genes in multiple species
- PMID: 35749360
- PMCID: PMC9245640
- DOI: 10.1073/pnas.2114441119
Convergent evolution of antiviral machinery derived from endogenous retrovirus truncated envelope genes in multiple species
Abstract
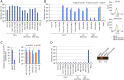
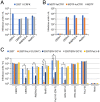
Host genetic resistance to viral infection controls the pathogenicity and epidemic dynamics of infectious diseases. Refrex-1 is a restriction factor against feline leukemia virus subgroup D (FeLV-D) and an endogenous retrovirus (ERV) in domestic cats (ERV-DC). Refrex-1 is encoded by a subset of ERV-DC loci with truncated envelope genes and secreted from cells as a soluble protein. Here, we identified the copper transporter CTR1 as the entry receptor for FeLV-D and genotype I ERV-DCs. We also identified CTR1 as a receptor for primate ERVs from crab-eating macaques and rhesus macaques, which were found in a search of intact envelope genes capable of forming infectious viruses. Refrex-1 counteracted infection by FeLV-D and ERV-DCs via competition for the entry receptor CTR1; the antiviral effects extended to primate ERVs with CTR1-dependent entry. Furthermore, truncated ERV envelope genes found in chimpanzee, bonobo, gorilla, crab-eating macaque, and rhesus macaque genomes could also block infection by feline and primate retroviruses. Genetic analyses showed that these ERV envelope genes were acquired in a species- or genus-specific manner during host evolution. These results indicated that soluble envelope proteins could suppress retroviral infection across species boundaries, suggesting that they function to control retroviral spread. Our findings revealed that several mammalian species acquired antiviral machinery from various ancient retroviruses, leading to convergent evolution for host defense.
Keywords: antiviral; evolution; receptor; restriction factor; retrovirus.
Conflict of interest statement
The authors declare no competing interests.
Figures


Similar articles
-
FeLIX is a restriction factor for mammalian retrovirus infection.J Virol. 2024 Apr 16;98(4):e0177123. doi: 10.1128/jvi.01771-23. Epub 2024 Mar 5. J Virol. 2024. PMID: 38440982 Free PMC article.
-
Identification of Copper Transporter 1 as a Receptor for Feline Endogenous Retrovirus ERV-DC14.J Virol. 2022 Jun 22;96(12):e0022922. doi: 10.1128/jvi.00229-22. Epub 2022 Jun 2. J Virol. 2022. PMID: 35652657 Free PMC article.
-
Tracking the Fate of Endogenous Retrovirus Segregation in Wild and Domestic Cats.J Virol. 2019 Nov 26;93(24):e01324-19. doi: 10.1128/JVI.01324-19. Print 2019 Dec 15. J Virol. 2019. PMID: 31534037 Free PMC article.
-
Tracking the Continuous Evolutionary Processes of an Endogenous Retrovirus of the Domestic Cat: ERV-DC.Viruses. 2018 Apr 6;10(4):179. doi: 10.3390/v10040179. Viruses. 2018. PMID: 29642384 Free PMC article. Review.
-
What role do endogenous retroviruses play in domestic cats infected with feline leukaemia virus?N Z Vet J. 2023 Jan;71(1):1-7. doi: 10.1080/00480169.2022.2131648. Epub 2022 Oct 26. N Z Vet J. 2023. PMID: 36178295 Review.
Cited by
-
Multiple recombination events between endogenous retroviral elements and feline leukemia virus.J Virol. 2024 Feb 20;98(2):e0140023. doi: 10.1128/jvi.01400-23. Epub 2024 Jan 19. J Virol. 2024. PMID: 38240589 Free PMC article.
-
Prevalence and genetic characterization of feline leukemia virus in portuguese stray cats.BMC Vet Res. 2025 Mar 26;21(1):205. doi: 10.1186/s12917-025-04691-2. BMC Vet Res. 2025. PMID: 40133929 Free PMC article.
-
Exploring the role of endogenous retroviruses in seasonal reproductive cycles: a case study of the ERV-V envelope gene in mink.Front Cell Infect Microbiol. 2024 Jun 28;14:1404431. doi: 10.3389/fcimb.2024.1404431. eCollection 2024. Front Cell Infect Microbiol. 2024. PMID: 39081866 Free PMC article.
-
FeLIX is a restriction factor for mammalian retrovirus infection.J Virol. 2024 Apr 16;98(4):e0177123. doi: 10.1128/jvi.01771-23. Epub 2024 Mar 5. J Virol. 2024. PMID: 38440982 Free PMC article.
-
Riboflavin transporter: evidence of a role as entry receptor for chimpanzee endogenous retrovirus.Virus Evol. 2025 May 7;11(1):veaf031. doi: 10.1093/ve/veaf031. eCollection 2025. Virus Evol. 2025. PMID: 40574751 Free PMC article.
References
-
- Belshaw R., Katzourakis A., Paces J., Burt A., Tristem M., High copy number in human endogenous retrovirus families is associated with copying mechanisms in addition to reinfection. Mol. Biol. Evol. 22, 814–817 (2005). - PubMed
-
- Spodick D. A., Ghosh A. K., Parimoo S., Roy-Burman P., The long terminal repeat of feline endogenous RD-114 retroviral DNAs: Analysis of transcription regulatory activity and nucleotide sequence. Virus Res. 9, 263–283 (1988). - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

