Adoptive Cellular Therapy with Autologous Tumor-Infiltrating Lymphocytes and T-cell Receptor-Engineered T Cells Targeting Common p53 Neoantigens in Human Solid Tumors
- PMID: 35749374
- PMCID: PMC9357191
- DOI: 10.1158/2326-6066.CIR-22-0040
Adoptive Cellular Therapy with Autologous Tumor-Infiltrating Lymphocytes and T-cell Receptor-Engineered T Cells Targeting Common p53 Neoantigens in Human Solid Tumors
Abstract
Adoptive cellular therapy (ACT) targeting neoantigens can achieve durable clinical responses in patients with cancer. Most neoantigens arise from patient-specific mutations, requiring highly individualized treatments. To broaden the applicability of ACT targeting neoantigens, we focused on TP53 mutations commonly shared across different cancer types. We performed whole-exome sequencing on 163 patients with metastatic solid cancers, identified 78 who had TP53 missense mutations, and through immunologic screening, identified 21 unique T-cell reactivities. Here, we report a library of 39 T-cell receptors (TCR) targeting TP53 mutations shared among 7.3% of patients with solid tumors. These TCRs recognized tumor cells in a TP53 mutation- and human leucocyte antigen (HLA)-specific manner in vitro and in vivo. Twelve patients with chemorefractory epithelial cancers were treated with ex vivo-expanded autologous tumor-infiltrating lymphocytes (TIL) that were naturally reactive against TP53 mutations. However, limited clinical responses (2 partial responses among 12 patients) were seen. These infusions contained low frequencies of mutant p53-reactive TILs that had exhausted phenotypes and showed poor persistence. We also treated one patient who had chemorefractory breast cancer with ACT comprising autologous peripheral blood lymphocytes transduced with an allogeneic HLA-A*02-restricted TCR specific for p53R175H. The infused cells exhibited an improved immunophenotype and prolonged persistence compared with TIL ACT and the patient experienced an objective tumor regression (-55%) that lasted 6 months. Collectively, these proof-of-concept data suggest that the library of TCRs targeting shared p53 neoantigens should be further evaluated for the treatment of patients with advanced human cancers. See related Spotlight by Klebanoff, p. 919.
©2022 The Authors; Published by the American Association for Cancer Research.
Figures
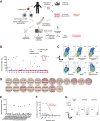
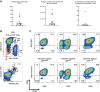
![Figure 2. Characterization of mutant p53–reactive TCRs. A, Tumor-cell recognition by p53Y220C-TCR (Y220C-TCR). Y220C-TCR was expressed in healthy donor PBLs and was tested against a panel of tumor cell lines with different TP53 mutations and HLAs. Following coculture with tumor cell lines, cell surface upregulation of 4–1BB on the T cells was measured by flow cytometry. Mock transduced T cells were included as a negative control (mean ± SEM, n = 3). B, Titration curve showing the avidity of Y220C-TCR against the WT and mutant Y220C ME. 4–1BB upregulation in healthy donor PBL transduced with Y220C-TCR following coculture with A*02:01+ T2 cells pulsed with the serially diluted ME was measured by flow cytometry (n = 1). C, Comparison of 4 HLA-A*02:01-restricted TCRs targeting p53R175H based on tumor cell reactivity. 4–1BB upregulation in p53R175H-TCR+CD8+ cells was measured by flow cytometry (mean ± SEM, n = 2). D, Titration curves for 4 TCRs targeting p53R175H. HLA-A*02:01+ T2 cells were pulsed with the serially diluted p53R175H ME and cocultured with TCR-transduced T cells. IFNγ secretion was measured by ELISA (mean ± SEM, n = 3). Statistical analysis by two-way ANOVA. Preclinical ACT of NSG mice bearing TYK-nu cancer cells using the R175H-TCR (4196-AV6/BV11)-engineered human PBL. Tumor size was calculated as the product of two perpendicular measurements. Tumor measurement was discontinued when the first mouse was euthanized [mean ± SEM, n = 4 (E and F), 5 (G, H, and J), and 10 (I)]. Donor information, transduction efficiency (TX; %), and CD8+ cell frequency (%) are given. E and G, Tumor growth following ACT of two different healthy donor PBLs transduced with the R175H-TCR or the irrelevant Y220C-TCR. I and J, Tumor growth following ACT of patient 4349’s PBL (1 or 2 × 107 cells) transduced with the R175H-TCR or untransduced (2 × 107 cells). J, Mice injected with either TYK-nu cells or the control 4259 PDX cells were treated with untransduced T cells, R175H-TCR–engineered T cells, or vehicle (PBS). Statistical analyses by two-way ANOVA (D, E, G, I, J) and by log-rank tests (F and H). *, P < 0.05; **, P < 0.01; ***, P < 0.001. A–D were independently repeated at least once.](https://cdn.ncbi.nlm.nih.gov/pmc/blobs/c0a2/9662943/cd71ff590a80/932fig2.gif)
Comment in
-
T-cell Receptor Gene Therapy Clinically Targeting a TP53 Public Neoantigen.Cancer Immunol Res. 2022 Aug 3;10(8):919. doi: 10.1158/2326-6066.CIR-22-0386. Cancer Immunol Res. 2022. PMID: 35767244
Comment on
-
T-cell Receptor Gene Therapy Clinically Targeting a TP53 Public Neoantigen.Cancer Immunol Res. 2022 Aug 3;10(8):919. doi: 10.1158/2326-6066.CIR-22-0386. Cancer Immunol Res. 2022. PMID: 35767244
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

