Chaperonin containing TCP1 as a marker for identification of circulating tumor cells in blood
- PMID: 35749519
- PMCID: PMC9232171
- DOI: 10.1371/journal.pone.0264651
Chaperonin containing TCP1 as a marker for identification of circulating tumor cells in blood
Abstract
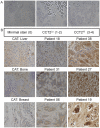
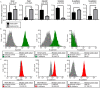
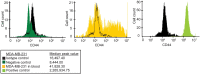
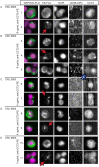
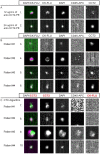
Herein we report the use of Chaperonin-Containing TCP-1 (CCT or TRiC) as a marker to detect circulating tumor cells (CTCs) that are shed from tumors during oncogenesis. Most detection methods used in liquid biopsy approaches for enumeration of CTCs from blood, employ epithelial markers like cytokeratin (CK). However, such markers provide little information on the potential of these shed tumor cells, which are normally short-lived, to seed metastatic sites. To identify a marker that could go beyond enumeration and provide actionable data on CTCs, we evaluated CCT. CCT is a protein-folding complex composed of eight subunits. Previously, we found that expression of the second subunit (CCT2 or CCTβ) inversely correlated with cancer patient survival and was essential for tumorigenesis in mice, driving tumor-promoting processes like proliferation and anchorage-independent growth. In this study, we examined CCT2 expression in cancer compared to normal tissues and found statistically significant increases in tumors. Because not all blood samples from cancer patients contain detectable CTCs, we used the approach of spiking a known number of cancer cells into blood from healthy donors to test a liquid biopsy approach using CCT2 to distinguish rare cancer cells from the large number of non-cancer cells in blood. Using a clinically validated method for capturing CTCs, we evaluated detection of intracellular CCT2 staining for visualization of breast cancer and small cell lung (SCLC) cancer cells. We demonstrated that CCT2 staining could be incorporated into a CTC capture and staining protocol, providing biologically relevant information to improve detection of cancer cells shed in blood. These results were confirmed with a pilot study of blood from SCLC patients. Our studies demonstrate that detection of CCT2 could identify rare cancer cells in blood and has application in liquid biopsy approaches to enhance the use of minimally invasive methods for cancer diagnosis.
Conflict of interest statement
We have read the journal’s policy and one of the authors of this manuscript (Dr. Annette Khaled) has the following competing interests: [shareholder in Seva Therapeutics, Inc.] This commercial entity holds a license to use intellectual property developed by the inventor (Dr. Khaled) and provided no funding and had no role in the design, preparation, or submission of this manuscript and did not employ any of the authors. This competing interest does not alter our adherence to PLOS ONE policies on sharing data and materials.
Figures


References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

