Genomics and pathotypes of the many faces of Escherichia coli
- PMID: 35749579
- PMCID: PMC9629502
- DOI: 10.1093/femsre/fuac031
Genomics and pathotypes of the many faces of Escherichia coli
Abstract
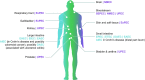
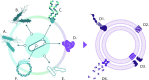
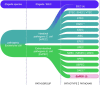
Escherichia coli is the most researched microbial organism in the world. Its varied impact on human health, consisting of commensalism, gastrointestinal disease, or extraintestinal pathologies, has generated a separation of the species into at least eleven pathotypes (also known as pathovars). These are broadly split into two groups, intestinal pathogenic E. coli (InPEC) and extraintestinal pathogenic E. coli (ExPEC). However, components of E. coli's infinite open accessory genome are horizontally transferred with substantial frequency, creating pathogenic hybrid strains that defy a clear pathotype designation. Here, we take a birds-eye view of the E. coli species, characterizing it from historical, clinical, and genetic perspectives. We examine the wide spectrum of human disease caused by E. coli, the genome content of the bacterium, and its propensity to acquire, exchange, and maintain antibiotic resistance genes and virulence traits. Our portrayal of the species also discusses elements that have shaped its overall population structure and summarizes the current state of vaccine development targeted at the most frequent E. coli pathovars. In our conclusions, we advocate streamlining efforts for clinical reporting of ExPEC, and emphasize the pathogenic potential that exists throughout the entire species.
Keywords: Escherichia coli pathotypes; accessory genome, antibiotic resistance; bacterial species; population dynamics; virulence factors.
© The Author(s) 2022. Published by Oxford University Press on behalf of FEMS.
Figures
References
-
- Abernethy JK, Johnson AP, Guy Ret al. Thirty day all-cause mortality in patients with Escherichia coli bacteraemia in England. Clin Microbiol Infect. 2015;21:251 e251–258. - PubMed
-
- Achtman M, Wagner M. Microbial diversity and the genetic nature of microbial species. Nat Rev Microbiol. 2008;6:431–40. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

