Advancing Precision Vaccinology by Molecular and Genomic Surveillance of Severe Acute Respiratory Syndrome Coronavirus 2 in Germany, 2021
- PMID: 35749674
- PMCID: PMC9278222
- DOI: 10.1093/cid/ciac399
Advancing Precision Vaccinology by Molecular and Genomic Surveillance of Severe Acute Respiratory Syndrome Coronavirus 2 in Germany, 2021
Abstract
Background: Comprehensive pathogen genomic surveillance represents a powerful tool to complement and advance precision vaccinology. The emergence of the Alpha variant in December 2020 and the resulting efforts to track the spread of this and other severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) variants of concern led to an expansion of genomic sequencing activities in Germany.
Methods: At Robert Koch Institute (RKI), the German National Institute of Public Health, we established the Integrated Molecular Surveillance for SARS-CoV-2 (IMS-SC2) network to perform SARS-CoV-2 genomic surveillance at the national scale, SARS-CoV-2-positive samples from laboratories distributed across Germany regularly undergo whole-genome sequencing at RKI.
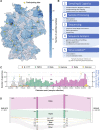
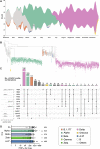
Results: We report analyses of 3623 SARS-CoV-2 genomes collected between December 2020 and December 2021, of which 3282 were randomly sampled. All variants of concern were identified in the sequenced sample set, at ratios equivalent to those in the 100-fold larger German GISAID sequence dataset from the same time period. Phylogenetic analysis confirmed variant assignments. Multiple mutations of concern emerged during the observation period. To model vaccine effectiveness in vitro, we employed authentic-virus neutralization assays, confirming that both the Beta and Zeta variants are capable of immune evasion. The IMS-SC2 sequence dataset facilitated an estimate of the SARS-CoV-2 incidence based on genetic evolution rates. Together with modeled vaccine efficacies, Delta-specific incidence estimation indicated that the German vaccination campaign contributed substantially to a deceleration of the nascent German Delta wave.
Conclusions: SARS-CoV-2 molecular and genomic surveillance may inform public health policies including vaccination strategies and enable a proactive approach to controlling coronavirus disease 2019 spread as the virus evolves.
Keywords: SARS-CoV-2; incidence estimation; molecular surveillance; pathogen genomics; variant of concern.
© The Author(s) 2022. Published by Oxford University Press on behalf of the Infectious Diseases Society of America.
Conflict of interest statement
Potential conflicts of interest. The authors report no potential conflicts of interest. All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
Figures


References
Publication types
MeSH terms
Supplementary concepts
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

