The international and intercontinental spread and expansion of antimicrobial-resistant Salmonella Typhi: a genomic epidemiology study
- PMID: 35750070
- PMCID: PMC9329132
- DOI: 10.1016/S2666-5247(22)00093-3
The international and intercontinental spread and expansion of antimicrobial-resistant Salmonella Typhi: a genomic epidemiology study
Abstract
Background: The emergence of increasingly antimicrobial-resistant Salmonella enterica serovar Typhi (S Typhi) threatens to undermine effective treatment and control. Understanding where antimicrobial resistance in S Typhi is emerging and spreading is crucial towards formulating effective control strategies.
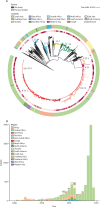
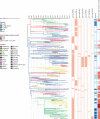
Methods: In this genomic epidemiology study, we sequenced the genomes of 3489 S Typhi strains isolated from prospective enteric fever surveillance studies in Nepal, Bangladesh, Pakistan, and India (between 2014 and 2019), and combined these with a global collection of 4169 S Typhi genome sequences isolated between 1905 and 2018 to investigate the temporal and geographical patterns of emergence and spread of antimicrobial-resistant S Typhi. We performed non-parametric phylodynamic analyses to characterise changes in the effective population size of fluoroquinolone-resistant, extensively drug-resistant (XDR), and azithromycin-resistant S Typhi over time. We inferred timed phylogenies for the major S Typhi sublineages and used ancestral state reconstruction methods to estimate the frequency and timing of international and intercontinental transfers.
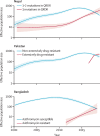
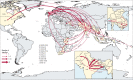
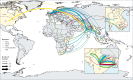
Findings: Our analysis revealed a declining trend of multidrug resistant typhoid in south Asia, except for Pakistan, where XDR S Typhi emerged in 2016 and rapidly replaced less-resistant strains. Mutations in the quinolone-resistance determining region (QRDR) of S Typhi have independently arisen and propagated on at least 94 occasions, nearly all occurring in south Asia. Strains with multiple QRDR mutations, including triple mutants with high-level fluoroquinolone resistance, have been increasing in frequency and displacing strains with fewer mutations. Strains containing acrB mutations, conferring azithromycin resistance, emerged in Bangladesh around 2013 and effective population size of these strains has been steadily increasing. We found evidence of frequent international (n=138) and intercontinental transfers (n=59) of antimicrobial-resistant S Typhi, followed by local expansion and replacement of drug-susceptible clades.
Interpretation: Independent acquisition of plasmids and homoplastic mutations conferring antimicrobial resistance have occurred repeatedly in multiple lineages of S Typhi, predominantly arising in south Asia before spreading to other regions.
Funding: Bill & Melinda Gates Foundation.
Copyright © 2022 The Author(s). Published by Elsevier Ltd. This is an Open Access article under the CC BY 4.0 license. Published by Elsevier Ltd.. All rights reserved.
Conflict of interest statement
Declaration of interests IB has consulted for BlueDot, a social benefit corporation that tracks the spread of emerging infectious diseases. SPL reports travel fees from the Bill & Melinda Gates Foundation. ZAD is a coordinator and founding member of the Global Typhoid Genomics Consortium and reports grants from EU Horizon 2020 for typhoid research outside the submitted work. All other authors declare no competing interests.
Figures
References
-
- Dougan G, Baker S. Salmonella enterica serovar Typhi and the pathogenesis of typhoid fever. Annu Rev Microbiol. 2014;68:317–336. - PubMed
-
- Mogasale V, Maskery B, Ochiai RL, et al. Burden of typhoid fever in low-income and middle-income countries: a systematic, literature-based update with risk-factor adjustment. Lancet Glob Health. 2014;2:e570–e580. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

