Gut microbiota combined with metabolites reveals unique features of acute myocardial infarction patients different from stable coronary artery disease
- PMID: 35750287
- PMCID: PMC10105070
- DOI: 10.1016/j.jare.2022.06.008
Gut microbiota combined with metabolites reveals unique features of acute myocardial infarction patients different from stable coronary artery disease
Abstract
Introduction: Acute myocardial infarction (AMI) accounts for the majority of deaths caused by coronary artery disease (CAD). Early warning of AMI, especially for patients with stable coronary artery disease (sCAD), is urgently needed. Our previous study showed that alterations in the gut microbiota were correlated with CAD severity.
Objectives: Herein, we tried to discover accurate and convenient biomarkers for AMI by combination of gut microbiota and fecal/blood/urinary metabolomics.
Methods: We recruited 190 volunteers including 93 sCAD patients, 49 AMI patients, and 48 subjects with normal coronary artery (NCA), and measured their blood biochemical parameters, 16S rRNA-based gut microbiota and NMR-based fecal/blood/urinary metabolites. We further selected 20 subjects from each group and analyzed their gut microbiota by whole-metagenome shotgun sequencing.
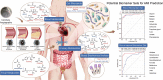
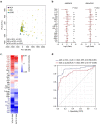
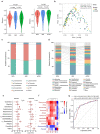
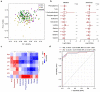
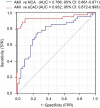
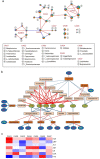
Results: Multi-omic analyses revealed that AMI patients exhibited specific changes in gut microbiota and serum/urinary/fecal metabolites as compared to subjects with sCAD or NCA. Fourteen bacterial genera and 30 metabolites (11 in feces, 10 in blood, 9 in urine) were closely related to AMI phenotypes and could accurately distinguish AMI patients from sCAD patients. Some species belonging to Alistipes, Streptococcus, Ruminococcus, Lactobacillus and Faecalibacterium were effective to distinguish AMI from sCAD and their predictive ability was confirmed in an independent cohort of CAD patients. We further selected nine indicators including 4 bacterial genera, 3 fecal and 2 urinary metabolites as a noninvasive biomarker set which can distinguish AMI from sCAD with an AUC of 0.932.
Conclusion: Combination of gut microbiota and fecal/urinary metabolites provided a set of potential useful and noninvasive predictive biomarker for AMI from sCAD.
Keywords: Acute myocardial infarction (AMI); Gut microbiota; Metabolite; Prediction model; Stable coronary artery disease (sCAD).
Copyright © 2023. Production and hosting by Elsevier B.V.
Conflict of interest statement
Declaration of Competing Interest The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Figures

References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

