Epigenetic regulation in cardiovascular disease: mechanisms and advances in clinical trials
- PMID: 35752619
- PMCID: PMC9233709
- DOI: 10.1038/s41392-022-01055-2
Epigenetic regulation in cardiovascular disease: mechanisms and advances in clinical trials
Abstract
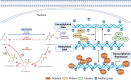
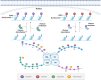
Epigenetics is closely related to cardiovascular diseases. Genome-wide linkage and association analyses and candidate gene approaches illustrate the multigenic complexity of cardiovascular disease. Several epigenetic mechanisms, such as DNA methylation, histone modification, and noncoding RNA, which are of importance for cardiovascular disease development and regression. Targeting epigenetic key enzymes, especially the DNA methyltransferases, histone methyltransferases, histone acetylases, histone deacetylases and their regulated target genes, could represent an attractive new route for the diagnosis and treatment of cardiovascular diseases. Herein, we summarize the knowledge on epigenetic history and essential regulatory mechanisms in cardiovascular diseases. Furthermore, we discuss the preclinical studies and drugs that are targeted these epigenetic key enzymes for cardiovascular diseases therapy. Finally, we conclude the clinical trials that are going to target some of these processes.
© 2022. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures





References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

