Assembly dynamics of FtsZ and DamX during infection-related filamentation and division in uropathogenic E. coli
- PMID: 35752634
- PMCID: PMC9233674
- DOI: 10.1038/s41467-022-31378-1
Assembly dynamics of FtsZ and DamX during infection-related filamentation and division in uropathogenic E. coli
Abstract
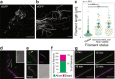
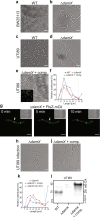
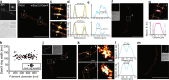
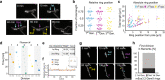
During infection of bladder epithelial cells, uropathogenic Escherichia coli (UPEC) can stop dividing and grow into highly filamentous forms. Here, we find that some filaments of E. coli UTI89 released from infected cells grow very rapidly and by more than 100 μm before initiating division, whereas others do not survive, suggesting that infection-related filamentation (IRF) is a stress response that promotes bacterial dispersal. IRF is accompanied by unstable, dynamic repositioning of FtsZ division rings. In contrast, DamX, which is associated with normal cell division and is also essential for IRF, is distributed uniformly around the cell envelope during filamentation. When filaments initiate division to regenerate rod cells, DamX condenses into stable rings prior to division. The DamX rings maintain consistent thickness during constriction and remain at the septum until after membrane fusion. Deletion of damX affects vegetative cell division in UTI89 (but not in the model E. coli K-12), and, during infection, blocks filamentation and reduces bacterial cell integrity. IRF therefore involves DamX distribution throughout the membrane and prevention of FtsZ ring stabilization, leading to cell division arrest. DamX then reassembles into stable division rings for filament division, promoting dispersal and survival during infection.
© 2022. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures



References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

