Designing optimized drug candidates with Generative Adversarial Network
- PMID: 35754029
- PMCID: PMC9233801
- DOI: 10.1186/s13321-022-00623-6
Designing optimized drug candidates with Generative Adversarial Network
Erratum in
-
Correction to: Designing optimized drug candidates with Generative Adversarial Network.J Cheminform. 2022 Aug 11;14(1):53. doi: 10.1186/s13321-022-00631-6. J Cheminform. 2022. PMID: 35953869 Free PMC article. No abstract available.
Abstract
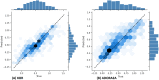
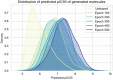
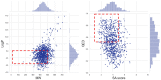
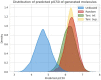
Drug design is an important area of study for pharmaceutical businesses. However, low efficacy, off-target delivery, time consumption, and high cost are challenges and can create barriers that impact this process. Deep Learning models are emerging as a promising solution to perform de novo drug design, i.e., to generate drug-like molecules tailored to specific needs. However, stereochemistry was not explicitly considered in the generated molecules, which is inevitable in targeted-oriented molecules. This paper proposes a framework based on Feedback Generative Adversarial Network (GAN) that includes optimization strategy by incorporating Encoder-Decoder, GAN, and Predictor deep models interconnected with a feedback loop. The Encoder-Decoder converts the string notations of molecules into latent space vectors, effectively creating a new type of molecular representation. At the same time, the GAN can learn and replicate the training data distribution and, therefore, generate new compounds. The feedback loop is designed to incorporate and evaluate the generated molecules according to the multiobjective desired property at every epoch of training to ensure a steady shift of the generated distribution towards the space of the targeted properties. Moreover, to develop a more precise set of molecules, we also incorporate a multiobjective optimization selection technique based on a non-dominated sorting genetic algorithm. The results demonstrate that the proposed framework can generate realistic, novel molecules that span the chemical space. The proposed Encoder-Decoder model correctly reconstructs 99% of the datasets, including stereochemical information. The model's ability to find uncharted regions of the chemical space was successfully shown by optimizing the unbiased GAN to generate molecules with a high binding affinity to the Kappa Opioid and Adenosine [Formula: see text] receptor. Furthermore, the generated compounds exhibit high internal and external diversity levels 0.88 and 0.94, respectively, and uniqueness.
Keywords: Drug design; GAN; Generative Adversial Network; Multiobjective optimization; NSGA; QSAR; RNN; SMILES.
© 2022. The Author(s).
Conflict of interest statement
The authors declare that they have no competing interests.
Figures

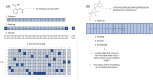
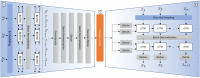
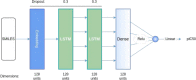

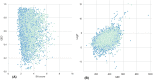


References
Grants and funding
- CENTRO-01-0145-FEDER-029266/Fundação para a Ciência e a Tecnologia
- CENTRO-01-0145-FEDER-029266/Fundação para a Ciência e a Tecnologia
- CENTRO-01-0145-FEDER-029266/Fundação para a Ciência e a Tecnologia
- CENTRO-01-0145-FEDER-029266/Fundação para a Ciência e a Tecnologia
- CENTRO-01-0145-FEDER-029266/Fundação para a Ciência e a Tecnologia
- CENTRO-01-0145-FEDER-029266/Fundação para a Ciência e a Tecnologia
- CENTRO-01-0145-FEDER-029266/Fundação para a Ciência e a Tecnologia
- CENTRO-01-0145-FEDER-029266/Fundação para a Ciência e a Tecnologia
- CENTRO-01-0145-FEDER-029266/Fundação para a Ciência e a Tecnologia
- CENTRO-01-0145-FEDER-029266/Fundação para a Ciência e a Tecnologia
LinkOut - more resources
Full Text Sources

