RNA Binding Protein Rbms1 Enables Neuronal Differentiation and Radial Migration during Neocortical Development by Binding and Stabilizing the RNA Message for Efr3a
- PMID: 35754370
- PMCID: PMC9385565
- DOI: 10.14348/molcells.2022.0044
RNA Binding Protein Rbms1 Enables Neuronal Differentiation and Radial Migration during Neocortical Development by Binding and Stabilizing the RNA Message for Efr3a
Abstract
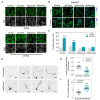
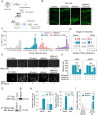
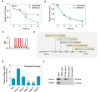
Various RNA-binding proteins (RBPs) are key components in RNA metabolism and contribute to several neurodevelop-mental disorders. To date, only a few of such RBPs have been characterized for their roles in neocortex development. Here, we show that the RBP, Rbms1, is required for radial migration, polarization and differentiation of neuronal progenitors to neurons in the neocortex development. Rbms1 expression is highest in the early development in the developing cortex, with its expression gradually diminishing from embryonic day 13.5 (E13.5) to postnatal day 0 (P0). From in utero electroporation (IUE) experiments when Rbms1 levels are knocked down in neuronal progenitors, their transition from multipolar to bipolar state is delayed and this is accompanied by a delay in radial migration of these cells. Reduced Rbms1 levels in vivo also reduces differentiation as evidenced by a decrease in levels of several differentiation markers, meanwhile having no significant effects on proliferation and cell cycle rates of these cells. As an RNA binding protein, we profiled the RNA binders of Rbms1 by a cross-linked-RIP sequencing assay, followed by quantitative real-time polymerase chain reaction verification and showed that Rbms1 binds and stabilizes the mRNA for Efr3a, a signaling adapter protein. We also demonstrate that ectopic Efr3a can recover the cells from the migration defects due to loss of Rbms1, both in vivo and in vitro migration assays with cultured cells. These imply that one of the functions of Rbms1 involves the stabilization of Efr3a RNA message, required for migration and maturation of neuronal progenitors in radial migration in the developing neocortex.
Keywords: Efr3a; RNA binding motif single stranded interacting protein 1; neurogenesis; radial migration.
Conflict of interest statement
The authors have no potential conflicts of interest to disclose.
Figures




References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

