A Prospective Review on Selectable Marker-Free Genome Engineered Rice: Past, Present and Future Scientific Realm
- PMID: 35754795
- PMCID: PMC9219106
- DOI: 10.3389/fgene.2022.882836
A Prospective Review on Selectable Marker-Free Genome Engineered Rice: Past, Present and Future Scientific Realm
Abstract
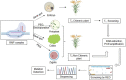
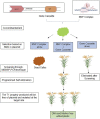
As a staple food crop, rice has gained mainstream attention in genome engineering for its genetic improvement. Genome engineering technologies such as transgenic and genome editing have enabled the significant improvement of target traits in relation to various biotic and abiotic aspects as well as nutrition, for which genetic diversity is lacking. In comparison to conventional breeding, genome engineering techniques are more precise and less time-consuming. However, one of the major issues with biotech rice commercialization is the utilization of selectable marker genes (SMGs) in the vector construct, which when incorporated into the genome are considered to pose risks to human health, the environment, and biodiversity, and thus become a matter of regulation. Various conventional strategies (co-transformation, transposon, recombinase systems, and MAT-vector) have been used in rice to avoid or remove the SMG from the developed events. However, the major limitations of these methods are; time-consuming, leftover cryptic sequences in the genome, and there is variable frequency. In contrast to these methods, CRISPR/Cas9-based marker excision, marker-free targeted gene insertion, programmed self-elimination, and RNP-based delivery enable us to generate marker-free engineered rice plants precisely and in less time. Although the CRISPR/Cas9-based SMG-free approaches are in their early stages, further research and their utilization in rice could help to break the regulatory barrier in its commercialization. In the current review, we have discussed the limitations of traditional methods followed by advanced techniques. We have also proposed a hypothesis, "DNA-free marker-less transformation" to overcome the regulatory barriers posed by SMGs.
Keywords: clustered regularly interspaced short palindromic repeats/crispr associated Cas9 (Crispr/Cas9); genetic engineering; genetically modified (GM) -regulation; rice; selectable marker genes (SMGs).
Copyright © 2022 Singh, Kaur, Praba, Kaur, Tanin, Kumar, Neelam, Sandhu and Vikal.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures


References
-
- Aliaga-Franco N., Zhang C., Presa S., Srivastava A. K., Granell A., Alabadí D., et al. (2019). Identification of Transgene-free CRISPR-Edited Plants of Rice, Tomato, and Arabidopsis by Monitoring DsRED Fluorescence in Dry Seeds. Front. Plant Sci. 10 (10), 1150. 10.3389/fpls.2019.01150 - DOI - PMC - PubMed
Publication types
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

