Identification and Expression Profiling of WRKY Family Genes in Sugarcane in Response to Bacterial Pathogen Infection and Nitrogen Implantation Dosage
- PMID: 35755708
- PMCID: PMC9218642
- DOI: 10.3389/fpls.2022.917953
Identification and Expression Profiling of WRKY Family Genes in Sugarcane in Response to Bacterial Pathogen Infection and Nitrogen Implantation Dosage
Abstract
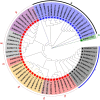
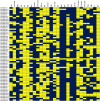
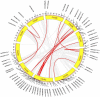
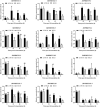
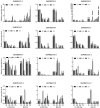
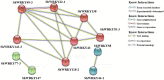
WRKY transcription factors (TFs) are essential players in different signaling cascades and regulatory networks involved in defense responses to various stressors. This study systematically analyzed and characterized WRKY family genes in the Saccharum spp. hybrid R570 and their expression in two sugarcane cultivars LCP85-384 (resistant to leaf scald) and ROC20 (susceptible to leaf scald) in response to bacterial pathogen infection and nitrogen implantation dosage. A total of 53 ShWRKY genes with 66 alleles were systematically identified in R570 based on the query sequence SsWRKY in S. spontaneum AP85-441. All ShRWKY alleles were further classified into four groups with 11 (16.7%) genes in group I, 36 (54.5%) genes in group II, 18 (27.3%) genes in group III, and 1 (1.5%) gene in group IV. Among them, 4 and 11 ShWRKY gene pairs displayed tandem and segmental duplication events, respectively. The ShWRKY genes exhibited conserved DNA-binding domains, which were accompanied by variations in introns, exons, and motifs. RT-qPCR analysis of two sugarcane cultivars triggered by Xanthomonas albilineans (Xa) revealed that four genes, ShWRKY13-2/39-1/49-3/125-3, exhibited significant upregulation in leaf scald-resistant LCP85-384. These WRKY genes were downregulated or unchanged in ROC20 at 24-72 h post-inoculation, suggesting that they play an important role in defense responses to Xa infection. Most of the 12 tested ShWRKYs, ShWRKY22-1/49-3/52-1 in particular, functioned as negative regulators in the two cultivars in response to a range of nitrogen (N) implantation doses. A total of 11 ShWRKY proteins were predicted to interact with each other. ShWRKY43 and ShWRKY49-3 are predicted to play core roles in the interaction network, as indicated by their interaction with six other ShWRKY proteins. Our results provide important candidate gene resources for the genetic improvement of sugarcane and lay the foundation for further functional characterization of ShWRKY genes in response to coupling effects of Xa infection and different N levels.
Keywords: Saccharum spp.; WRKY transcription factors; Xanthomonas albilineans; gene regulation; nitrogen dosage.
Copyright © 2022 Javed, Zhou, Li, Hu, Wang and Gao.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures

References
LinkOut - more resources
Full Text Sources
Miscellaneous

