Metagenomic Screening for Lipolytic Genes Reveals an Ecology-Clustered Distribution Pattern
- PMID: 35756004
- PMCID: PMC9226776
- DOI: 10.3389/fmicb.2022.851969
Metagenomic Screening for Lipolytic Genes Reveals an Ecology-Clustered Distribution Pattern
Abstract
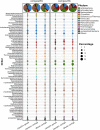
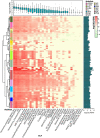
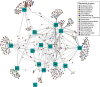
Lipolytic enzymes are one of the most important enzyme types for application in various industrial processes. Despite the continuously increasing demand, only a small portion of the so far encountered lipolytic enzymes exhibit adequate stability and activities for biotechnological applications. To explore novel and/or extremophilic lipolytic enzymes, microbial consortia in two composts at thermophilic stage were analyzed using function-driven and sequence-based metagenomic approaches. Analysis of community composition by amplicon-based 16S rRNA genes and transcripts, and direct metagenome sequencing revealed that the communities of the compost samples were dominated by members of the phyla Actinobacteria, Proteobacteria, Firmicutes, Bacteroidetes, and Chloroflexi. Function-driven screening of the metagenomic libraries constructed from the two samples yielded 115 unique lipolytic enzymes. The family assignment of these enzymes was conducted by analyzing the phylogenetic relationship and generation of a protein sequence similarity network according to an integrated classification system. The sequence-based screening was performed by using a newly developed database, containing a set of profile Hidden Markov models, highly sensitive and specific for detection of lipolytic enzymes. By comparing the lipolytic enzymes identified through both approaches, we demonstrated that the activity-directed complements sequence-based detection, and vice versa. The sequence-based comparative analysis of lipolytic genes regarding diversity, function and taxonomic origin derived from 175 metagenomes indicated significant differences between habitats. Analysis of the prevalent and distinct microbial groups providing the lipolytic genes revealed characteristic patterns and groups driven by ecological factors. The here presented data suggests that the diversity and distribution of lipolytic genes in metagenomes of various habitats are largely constrained by ecological factors.
Keywords: comparative analysis; compost; function-driven metagenomics; lipolytic enzyme classification; lipolytic enzymes; profile HMM; sequence-based metagenomics.
Copyright © 2022 Lu, Schneider and Daniel.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures


Similar articles
-
The combination of functional metagenomics and an oil-fed enrichment strategy revealed the phylogenetic diversity of lipolytic bacteria overlooked by the cultivation-based method.Microbes Environ. 2014;29(2):154-61. doi: 10.1264/jsme2.me14002. Epub 2014 May 23. Microbes Environ. 2014. PMID: 24859309 Free PMC article.
-
Metagenomic analysis of microbial consortia enriched from compost: new insights into the role of Actinobacteria in lignocellulose decomposition.Biotechnol Biofuels. 2016 Jan 29;9:22. doi: 10.1186/s13068-016-0440-2. eCollection 2016. Biotechnol Biofuels. 2016. PMID: 26834834 Free PMC article.
-
Microbial diversity and biochemical potential encoded by thermal spring metagenomes derived from the Kamchatka Peninsula.Archaea. 2013;2013:136714. doi: 10.1155/2013/136714. Epub 2013 Feb 27. Archaea. 2013. PMID: 23533327 Free PMC article.
-
Metagenomics: Probing pollutant fate in natural and engineered ecosystems.Biotechnol Adv. 2016 Dec;34(8):1413-1426. doi: 10.1016/j.biotechadv.2016.10.006. Epub 2016 Nov 5. Biotechnol Adv. 2016. PMID: 27825829 Review.
-
Practical considerations for sampling and data analysis in contemporary metagenomics-based environmental studies.J Microbiol Methods. 2018 Nov;154:14-18. doi: 10.1016/j.mimet.2018.09.020. Epub 2018 Oct 1. J Microbiol Methods. 2018. PMID: 30287354 Review.
Cited by
-
Engineered polyethylene terephthalate hydrolases: perspectives and limits.Appl Microbiol Biotechnol. 2024 Jul 2;108(1):404. doi: 10.1007/s00253-024-13222-2. Appl Microbiol Biotechnol. 2024. PMID: 38953996 Free PMC article. Review.
References
-
- Alisch M., Feuerhack A., Müller H., Mensak B., Andreaus J., Zimmermann W. (2004). Biocatalytic modification of polyethylene terephthalate fibres by esterases from actinomycete isolates. Biocatal. Biotransform. 22 347–351. 10.1080/10242420400025877 - DOI
-
- Andrews S. (2010). FastQC: A Quality Control Tool for High Throughput Sequence Data. Available online at: http://www.bioinformatics.babraham.ac.uk/projects/fastqc/
LinkOut - more resources
Full Text Sources

