Clustered Regularly Interspaced Short Palindromic Repeat/Cas12a Mediated Multiplexable and Portable Detection Platform for GII Genotype Porcine Epidemic Diarrhoea Virus Rapid Diagnosis
- PMID: 35756009
- PMCID: PMC9218691
- DOI: 10.3389/fmicb.2022.920801
Clustered Regularly Interspaced Short Palindromic Repeat/Cas12a Mediated Multiplexable and Portable Detection Platform for GII Genotype Porcine Epidemic Diarrhoea Virus Rapid Diagnosis
Abstract
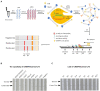
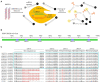
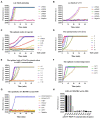
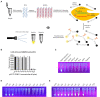
Porcine epidemic diarrhoea virus (PEDV) is a member of the genus Alphacoronavirus in the family Coronaviridae. It causes acute watery diarrhoea and vomiting in piglets with high a mortality rate. Currently, the GII genotype, PEDV, possesses a high separation rate in wild strains and is usually reported in immunity failure cases, which indicates a need for a portable and sensitive detection method. Here, reverse transcription-recombinase aided amplification (RT-RAA) was combined with the Clustered Regularly Interspaced Short Palindromic Repeat (CRISPR)/Cas12a system to establish a multiplexable, rapid and portable detection platform for PEDV. The CRISPR RNA (crRNA) against Spike (S) gene of GII PEDV specifically were added into the protocol. This system is suitable for different experimental conditions, including ultra-sensitive fluorescence, visual, UV light, or flow strip detection. Moreover, it exhibits high sensitivity and specificity and can detect at least 100 copies of the target gene in each reaction. The CRISPR/Cas12a detection platform requires less time and represents a rapid, reliable and practical tool for the rapid diagnosis of GII genotype PEDV.
Keywords: CRISPR/Cas12a; GII genotype; lateral flow strip; porcine epidemic diarrhoea virus; rapid diagnosis.
Copyright © 2022 Qian, Liao, Zeng, Peng, Wu, Li, Bo, Hu, Nan, Wen, Cao, Xue, Zhang and Dai.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures
References
-
- Chen Q., Li G., Stasko J., Thomas J. T., Stensland W. R., Pillatzki A. E., et al. (2014). Isolation and characterization of porcine epidemic diarrhea viruses associated with the 2013 disease outbreak among swine in the United States. J. Clin. Microbiol. 52, 234–243. doi: 10.1128/JCM.02820-13, PMID: - DOI - PMC - PubMed
Associated data
LinkOut - more resources
Full Text Sources

