Bacteriophage-Resistant Mutant of Enterococcus faecalis Is Impaired in Biofilm Formation
- PMID: 35756031
- PMCID: PMC9218719
- DOI: 10.3389/fmicb.2022.913023
Bacteriophage-Resistant Mutant of Enterococcus faecalis Is Impaired in Biofilm Formation
Abstract
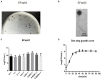
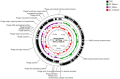
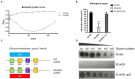
Enterococcus faecalis is a common gram-positive non-spore-forming bacterium in nature and is found in the upper respiratory tract, intestine, and mouth of healthy people. E. faecalis is also one of the common pathogens causing nosocomial infections and is resistant to several antibiotics commonly used in practice. Thus, treating drug-resistant E. faecalis with antibiotics is challenging, and new approaches are needed. In this study, we isolated a bacteriophage named EFap02 that targets E. faecalis strain EFa02 from sewage at Southwest Hospital. Phage EFap02 belongs to the Siphoviridae family with a long tail of approximately 210 nm, and EFap02 can tolerate a strong acid and alkali environment and high temperature. Its receptor was identified as the capsular polysaccharide. Phage-resistant mutants had loss-of-function mutations in glycosyltransferase (gtr2), which is responsible for capsular polysaccharide biosynthesis, and this caused the loss of capsular polysaccharide and interruption of phage adsorption. Although phage-resistant mutants against EFap02 can be selected, such mutants are impaired in biofilm formation due to the loss of capsular polysaccharide, which compromises its virulence. Therefore, this study provided a detailed description of the E. faecalis EFap02 phage with the potential for treating E. faecalis infection.
Keywords: Enterococcus faecalis; bacteriophage; biofilm; capsular polysaccharide; phage receptor; phage resistance.
Copyright © 2022 Liu, Zhu, Li, Lu, Xiong, Zhong and Wang.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures
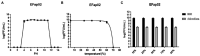


Similar articles
-
Enterococcus faecalis Bacteriophage vB_EfaS_efap05-1 Targets the Surface Polysaccharide and ComEA Protein as the Receptors.Front Microbiol. 2022 Mar 31;13:866382. doi: 10.3389/fmicb.2022.866382. eCollection 2022. Front Microbiol. 2022. PMID: 35432223 Free PMC article.
-
Anti-biofilm Activity of a Lytic Phage Against Vancomycin-Resistant Enterococcus faecalis.Iran J Pathol. 2022 Summer;17(3):285-293. doi: 10.30699/IJP.2022.541855.2760. Epub 2022 Aug 12. Iran J Pathol. 2022. PMID: 36247507 Free PMC article.
-
Molecular Basis for Lytic Bacteriophage Resistance in Enterococci.mBio. 2016 Aug 30;7(4):e01304-16. doi: 10.1128/mBio.01304-16. mBio. 2016. PMID: 27578757 Free PMC article.
-
Phage therapy against Enterococcus faecalis in dental root canals.J Oral Microbiol. 2016 Sep 16;8:32157. doi: 10.3402/jom.v8.32157. eCollection 2016. J Oral Microbiol. 2016. PMID: 27640530 Free PMC article. Review.
-
The Regulations of Essential WalRK Two-Component System on Enterococcus faecalis.J Clin Med. 2023 Jan 18;12(3):767. doi: 10.3390/jcm12030767. J Clin Med. 2023. PMID: 36769415 Free PMC article. Review.
Cited by
-
Enterococcal Phages: Food and Health Applications.Antibiotics (Basel). 2023 May 2;12(5):842. doi: 10.3390/antibiotics12050842. Antibiotics (Basel). 2023. PMID: 37237745 Free PMC article. Review.
-
Virulent Phage vB_EfaS_WH1 Removes Enterococcus faecalis Biofilm and Inhibits Its Growth on the Surface of Chicken Meat.Viruses. 2023 May 20;15(5):1208. doi: 10.3390/v15051208. Viruses. 2023. PMID: 37243294 Free PMC article.
-
Autographiviridae phage HH109 uses capsular polysaccharide for infection of Vibrio alginolyticus.iScience. 2024 Aug 8;27(9):110695. doi: 10.1016/j.isci.2024.110695. eCollection 2024 Sep 20. iScience. 2024. PMID: 39252973 Free PMC article.
-
Horizontal Gene Transfer of Antibiotic Resistance Genes in Biofilms.Antibiotics (Basel). 2023 Feb 4;12(2):328. doi: 10.3390/antibiotics12020328. Antibiotics (Basel). 2023. PMID: 36830238 Free PMC article. Review.
-
Investigating the phenotypic alterations associated with hypermucoviscous hypervirulent Klebsiella pneumoniae during phage resistance development.BMC Microbiol. 2025 Aug 22;25(1):528. doi: 10.1186/s12866-025-04268-x. BMC Microbiol. 2025. PMID: 40841996 Free PMC article.
References
-
- Bao J., Wu N., Zeng Y., Chen L., Li L., Yang L., et al. . (2020). Non-active antibiotic and bacteriophage synergism to successfully treat recurrent urinary tract infection caused by extensively drug-resistant. Emerg. Microbes Infect. 9, 771–774. doi: 10.1080/22221751.2020.1747950, PMID: - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources

