Quantifying DNA damage on paper sensors via controlled template-independent DNA polymerization
- PMID: 35756503
- PMCID: PMC9172109
- DOI: 10.1039/d1sc04268h
Quantifying DNA damage on paper sensors via controlled template-independent DNA polymerization
Abstract
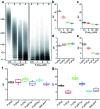
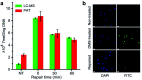
We report on a paper-based sensor capable of performing template-independent DNA synthesis by terminal deoxynucleotidyl transferase (TdT). Importantly, we observed that TdT efficiently incorporates fluorescently labeled dUTP on to 3'-OH ends of DNA strands in a strictly controllable manner on cellulose paper, in comparison to its distributive mode of DNA synthesis in solution. Due to the high roughness and porous nature of cellulose paper, we attribute this controllable DNA polymerization to the pore confinement effect on the catalytic behaviour of TdT. Taking advantage of this finding, we proposed a paper-assisted TdT (PAT) assay for absolute quantification of alkylated DNA lesions (N7-methylguanine), DNA deamination (cytosine-to-uracil) and DNA oxidation (8-oxo-7,8-dihydroguanine) by combining various DNA glycosylases. This PAT assay provides a low-cost, high throughput and easy to use method for quantifying the absolute levels of various types of DNA lesions, thus making it well-suited for drug development, genotoxicity testing, and environmental toxicology.
This journal is © The Royal Society of Chemistry.
Conflict of interest statement
There are no conflicts to declare.
Figures




References
-
- Martinez A. W. Phillips S. T. Whitesides G. M. Anal. Chem. 2010;82:3. doi: 10.1021/ac9013989. - DOI - PubMed
- Parolo C. Merkoçi A. Chem. Soc. Rev. 2013;42:450. doi: 10.1039/C2CS35255A. - DOI - PubMed
- Gong M. M. Sinton D. Chem. Rev. 2017;117:8447. doi: 10.1021/acs.chemrev.7b00024. - DOI - PubMed
- Yang Y. Y. Noviana E. Nguyen M. P. Geiss B. J. Dandy D. S. Henry C. S. Anal. Chem. 2017;89:71. doi: 10.1021/acs.analchem.6b04581. - DOI - PubMed
- Zhang Y. Zhang Q. Cheng F. Chang Y. Liu M. Li Y. Chem. Sci. 2021;12:8282. doi: 10.1039/D1SC00795E. - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources

