The Chromatin Landscape Channels DNA Double-Strand Breaks to Distinct Repair Pathways
- PMID: 35757003
- PMCID: PMC9213757
- DOI: 10.3389/fcell.2022.909696
The Chromatin Landscape Channels DNA Double-Strand Breaks to Distinct Repair Pathways
Abstract
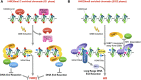
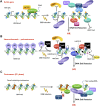
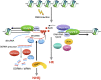
DNA double-strand breaks (DSBs), the most deleterious DNA lesions, are primarily repaired by two pathways, namely homologous recombination (HR) and non-homologous end joining (NHEJ), the choice of which is largely dependent on cell cycle phase and the local chromatin landscape. Recent studies have revealed that post-translational modifications on histones play pivotal roles in regulating DSB repair pathways including repair pathway choice. In this review, we present our current understanding of how these DSB repair pathways are employed in various chromatin landscapes to safeguard genomic integrity. We place an emphasis on the impact of different histone post-translational modifications, characteristic of euchromatin or heterochromatin regions, on DSB repair pathway choice. We discuss the potential roles of damage-induced chromatin modifications in the maintenance of genome and epigenome integrity. Finally, we discuss how RNA transcripts from the vicinity of DSBs at actively transcribed regions also regulate DSB repair pathway choice.
Keywords: DNA doube-strand breaks; RNA-DNA hybrids; euchromatin; heterochromatin; histone modifications; repair pathway choice; transcription.
Copyright © 2022 Chen and Tyler.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures
Similar articles
-
The Chromatin Landscape around DNA Double-Strand Breaks in Yeast and Its Influence on DNA Repair Pathway Choice.Int J Mol Sci. 2023 Feb 7;24(4):3248. doi: 10.3390/ijms24043248. Int J Mol Sci. 2023. PMID: 36834658 Free PMC article. Review.
-
Timely double-strand break repair and pathway choice in pericentromeric heterochromatin depend on the histone demethylase dKDM4A.Genes Dev. 2019 Jan 1;33(1-2):103-115. doi: 10.1101/gad.317537.118. Epub 2018 Dec 21. Genes Dev. 2019. PMID: 30578303 Free PMC article.
-
DNA double strand break repair pathway choice: a chromatin based decision?Nucleus. 2015;6(2):107-13. doi: 10.1080/19491034.2015.1010946. Epub 2015 Feb 12. Nucleus. 2015. PMID: 25675367 Free PMC article.
-
Histone acetylation dynamics in repair of DNA double-strand breaks.Front Genet. 2022 Sep 9;13:926577. doi: 10.3389/fgene.2022.926577. eCollection 2022. Front Genet. 2022. PMID: 36159966 Free PMC article. Review.
-
The influence of heterochromatin on DNA double strand break repair: Getting the strong, silent type to relax.DNA Repair (Amst). 2010 Dec 10;9(12):1273-82. doi: 10.1016/j.dnarep.2010.09.013. Epub 2010 Oct 30. DNA Repair (Amst). 2010. PMID: 21036673 Review.
Cited by
-
Male-biased recombination at chromosome ends in a songbird revealed by precisely mapping crossover positions.G3 (Bethesda). 2024 Sep 4;14(9):jkae150. doi: 10.1093/g3journal/jkae150. G3 (Bethesda). 2024. PMID: 38985659 Free PMC article.
-
From Double-Strand Break Recognition to Cell-Cycle Checkpoint Activation: High Content and Resolution Image Cytometry Unmasks 53BP1 Multiple Roles in DNA Damage Response and p53 Action.Int J Mol Sci. 2022 Sep 5;23(17):10193. doi: 10.3390/ijms231710193. Int J Mol Sci. 2022. PMID: 36077590 Free PMC article.
-
Automated Image Analysis of Transmission Electron Micrographs: Nanoscale Evaluation of Radiation-Induced DNA Damage in the Context of Chromatin.Cells. 2023 Oct 10;12(20):2427. doi: 10.3390/cells12202427. Cells. 2023. PMID: 37887271 Free PMC article.
-
NuRD chromatin remodeling is required to repair exogenous DSBs in the Caenorhabditis elegans germline.bioRxiv [Preprint]. 2024 Sep 15:2024.09.14.613027. doi: 10.1101/2024.09.14.613027. bioRxiv. 2024. PMID: 39314477 Free PMC article. Preprint.
-
DNA Damage, Genome Stability, and Adaptation: A Question of Chance or Necessity?Genes (Basel). 2024 Apr 21;15(4):520. doi: 10.3390/genes15040520. Genes (Basel). 2024. PMID: 38674454 Free PMC article. Review.
References
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources

