Inhibition of the Protein Arginine Methyltransferase PRMT5 in High-Risk Multiple Myeloma as a Novel Treatment Approach
- PMID: 35757005
- PMCID: PMC9213887
- DOI: 10.3389/fcell.2022.879057
Inhibition of the Protein Arginine Methyltransferase PRMT5 in High-Risk Multiple Myeloma as a Novel Treatment Approach
Abstract
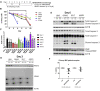
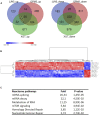
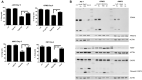
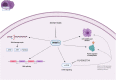
Multiple myeloma (MM) is an incurable clonal plasma cell malignancy. Subsets of patients have high-risk features linked with dismal outcome. Therefore, the need for effective therapeutic options remains high. Here, we used bio-informatic tools to identify novel targets involved in DNA repair and epigenetics and which are associated with high-risk myeloma. The prognostic significance of the target genes was analyzed using publicly available gene expression data of MM patients (TT2/3 and HM cohorts). Hence, protein arginine methyltransferase 5 (PRMT5) was identified as a promising target. Druggability was assessed in OPM2, JJN3, AMO1 and XG7 human myeloma cell lines using the PRMT5-inhibitor EPZ015938. EPZ015938 strongly reduced the total symmetric-dimethyl arginine levels in all cell lines and lead to decreased cellular growth, supported by cell line dependent changes in cell cycle distribution. At later time points, apoptosis occurred, as evidenced by increased AnnexinV-positivity and cleavage of PARP and caspases. Transcriptome analysis revealed a role for PRMT5 in regulating alternative splicing, nonsense-mediated decay, DNA repair and PI3K/mTOR-signaling, irrespective of the cell line type. PRMT5 inhibition reduced the expression of upstream DNA repair kinases ATM and ATR, which may in part explain our observation that EPZ015938 and the DNA-alkylating agent, melphalan, have combinatory effects. Of interest, using a low-dose of mTOR-inhibitor, we observed that cell viability was partially rescued from the effects of EPZ015938, indicating a role for mTOR-related pathways in the anti-myeloma activity of EPZ015938. Moreover, PRMT5 was shown to be involved in splicing regulation of MMSET and SLAMF7, known genes of importance in MM disease. As such, we broaden the understanding of the exact role of PRMT5 in MM disease and further underline its use as a possible therapeutic target.
Keywords: DNA repair; PRMT5; RNA splicing; epigenetics; myeloma.
Copyright © 2022 Vlummens, Verhulst, De Veirman, Maes, Menu, Moreaux, De Boussac, Robert, De Bruyne, Hose, Offner, Vanderkerken and Maes.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures


Similar articles
-
PRMT5 Modulates Splicing for Genome Integrity and Preserves Proteostasis of Hematopoietic Stem Cells.Cell Rep. 2019 Feb 26;26(9):2316-2328.e6. doi: 10.1016/j.celrep.2019.02.001. Cell Rep. 2019. PMID: 30811983
-
SAM-Competitive PRMT5 Inhibitor PF-06939999 Demonstrates Antitumor Activity in Splicing Dysregulated NSCLC with Decreased Liability of Drug Resistance.Mol Cancer Ther. 2022 Jan;21(1):3-15. doi: 10.1158/1535-7163.MCT-21-0620. Epub 2021 Nov 4. Mol Cancer Ther. 2022. PMID: 34737197
-
The protein arginine methyltransferase PRMT5 confers therapeutic resistance to mTOR inhibition in glioblastoma.J Neurooncol. 2019 Oct;145(1):11-22. doi: 10.1007/s11060-019-03274-0. Epub 2019 Aug 31. J Neurooncol. 2019. PMID: 31473880 Free PMC article.
-
Protein arginine N-methyltransferase 5 in colorectal carcinoma: Insights into mechanisms of pathogenesis and therapeutic strategies.Biomed Pharmacother. 2022 Jan;145:112368. doi: 10.1016/j.biopha.2021.112368. Epub 2021 Nov 15. Biomed Pharmacother. 2022. PMID: 34794114 Review.
-
PRMT5: An Emerging Target for Pancreatic Adenocarcinoma.Cancers (Basel). 2021 Oct 13;13(20):5136. doi: 10.3390/cancers13205136. Cancers (Basel). 2021. PMID: 34680285 Free PMC article. Review.
Cited by
-
Inhibiting PRMT5 induces DNA damage and increases anti-proliferative activity of Niraparib, a PARP inhibitor, in models of breast and ovarian cancer.BMC Cancer. 2023 Aug 18;23(1):775. doi: 10.1186/s12885-023-11260-z. BMC Cancer. 2023. PMID: 37596538 Free PMC article.
-
PRMT blockade induces defective DNA replication stress response and synergizes with PARP inhibition.Cell Rep Med. 2023 Dec 19;4(12):101326. doi: 10.1016/j.xcrm.2023.101326. Cell Rep Med. 2023. PMID: 38118413 Free PMC article.
-
Emerging roles of protein arginine methyltransferase in multiple myeloma.Mol Ther Oncol. 2025 May 26;33(2):201003. doi: 10.1016/j.omton.2025.201003. eCollection 2025 Jun 18. Mol Ther Oncol. 2025. PMID: 40524859 Free PMC article. Review.
References
-
- Barlogie B., Tricot G., Rasmussen E., Anaissie E., van Rhee F., Zangari M., et al. (2006). Total Therapy 2 without Thalidomide in Comparison with Total Therapy 1: Role of Intensified Induction and Posttransplantation Consolidation Therapies. Blood 7(7), 2633–2638. 10.1182/blood-2005-10-4084 - DOI - PubMed
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

