Multiomic Analysis of the Gut Microbiome in Psoriasis Reveals Distinct Host‒Microbe Associations
- PMID: 35757783
- PMCID: PMC9214347
- DOI: 10.1016/j.xjidi.2022.100115
Multiomic Analysis of the Gut Microbiome in Psoriasis Reveals Distinct Host‒Microbe Associations
Abstract
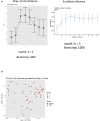
Psoriasis is a chronic, inflammatory skin disease that affects 2‒3% of the global population. Besides skin manifestations, patients with psoriasis have increased susceptibility to a number of comorbidities, including psoriatic arthritis, cardiovascular disease, and inflammatory bowel disease. To understand the systemic component of psoriasis pathogenesis, we performed a pilot study to examine the fecal metagenome, host colonic transcriptome, and host peripheral blood immune profiles of patients with psoriasis and healthy controls. Our study showed increased functional diversity in the gut microbiome of patients with psoriasis. In addition, we identified microbial species that preferentially associate with patients with psoriasis and which have been previously found to associate with other autoimmune diseases. Intriguingly, our data revealed three psoriasis subgroups that have distinct microbial and host features. Integrating these features revealed host‒microbe associations that are specific to psoriasis or particular psoriasis subgroups. Our findings provide insight into the factors that may affect the development of comorbidities in patients with psoriasis and may hold diagnostic potential for early identification of patients with psoriasis at risk for these comorbidities.
Keywords: IBD, inflammatory bowel disease; RNA-seq, RNA sequencing.
© 2022 The Authors.
Figures














References
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

