Epigenetic rewiring of pathways related to odour perception in immune cells exposed to SARS-CoV-2 in vivo and in vitro
- PMID: 35758003
- PMCID: PMC9665140
- DOI: 10.1080/15592294.2022.2089471
Epigenetic rewiring of pathways related to odour perception in immune cells exposed to SARS-CoV-2 in vivo and in vitro
Abstract
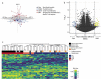
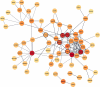
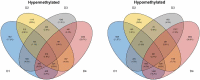
A majority of SARS-CoV-2 recoverees develop only mild-to-moderate symptoms, while some remain completely asymptomatic. Although viruses, including SARS-CoV-2, may evade host immune responses by epigenetic mechanisms including DNA methylation, little is known about whether these modifications are important in defence against and healthy recovery from COVID-19 in the host. To this end, epigenome-wide DNA methylation patterns from COVID-19 convalescents were compared to uninfected controls from before and after the pandemic. Peripheral blood mononuclear cell (PBMC) DNA was extracted from uninfected controls, COVID-19 convalescents, and symptom-free individuals with SARS-CoV-2-specific T cell-responses, as well as from PBMCs stimulated in vitro with SARS-CoV-2. Subsequently, the Illumina MethylationEPIC 850K array was performed, and statistical/bioinformatic analyses comprised differential DNA methylation, pathway over-representation, and module identification analyses. Differential DNA methylation patterns distinguished COVID-19 convalescents from uninfected controls, with similar results in an experimental SARS-CoV-2 infection model. A SARS-CoV-2-induced module was identified in vivo, comprising 66 genes of which six (TP53, INS, HSPA4, SP1, ESR1, and FAS) were present in corresponding in vitro analyses. Over-representation analyses revealed involvement in Wnt, muscarinic acetylcholine receptor signalling, and gonadotropin-releasing hormone receptor pathways. Furthermore, numerous differentially methylated and network genes from both settings interacted with the SARS-CoV-2 interactome. Altered DNA methylation patterns of COVID-19 convalescents suggest recovery from mild-to-moderate SARS-CoV-2 infection leaves longstanding epigenetic traces. Both in vitro and in vivo exposure caused epigenetic modulation of pathways thataffect odour perception. Future studies should determine whether this reflects host-induced protective antiviral defense or targeted viral hijacking to evade host defence.
Keywords: DNA methylation; PBMC; SARS-CoV-2; in vitro stimulation; interactome; mild-to-moderate; module identification; network analysis.
Conflict of interest statement
M.L., S.Sa., and J.D. have prepared and filed a patent based on the findings from the present study. None of the remaining authors declare any competing interests.
Figures

References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous
