Vfold-Pipeline: a web server for RNA 3D structure prediction from sequences
- PMID: 35758624
- PMCID: PMC9364377
- DOI: 10.1093/bioinformatics/btac426
Vfold-Pipeline: a web server for RNA 3D structure prediction from sequences
Abstract
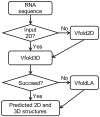
Summary: RNA 3D structures are critical for understanding their functions and for RNA-targeted drug design. However, experimental determination of RNA 3D structures is laborious and technically challenging, leading to the huge gap between the number of sequences and the availability of RNA structures. Therefore, the computer-aided structure prediction of RNA 3D structures from sequences becomes a highly desirable solution to this problem. Here, we present a pipeline server for RNA 3D structure prediction from sequences that integrates the Vfold2D, Vfold3D and VfoldLA programs. The Vfold2D program can incorporate the SHAPE experimental data in 2D structure prediction. The pipeline can also automatically extract 2D structural constraints from the Rfam database. Furthermore, with a significantly expanded 3D template database for various motifs, this Vfold-Pipeline server can efficiently return accurate 3D structure predictions or reliable initial 3D structures for further refinement.
Availability and implementation: http://rna.physics.missouri.edu/vfoldPipeline/index.html. The data underlying this article have been provided in the article and in its online supplementary material.
Supplementary information: Supplementary data are available at Bioinformatics online.
© The Author(s) 2022. Published by Oxford University Press. All rights reserved. For permissions, please e-mail: journals.permissions@oup.com.
Figures
Similar articles
-
VfoldLA: A web server for loop assembly-based prediction of putative 3D RNA structures.J Struct Biol. 2019 Sep 1;207(3):235-240. doi: 10.1016/j.jsb.2019.06.002. Epub 2019 Jun 4. J Struct Biol. 2019. PMID: 31173857 Free PMC article.
-
Predicting RNA Scaffolds with a Hybrid Method of Vfold3D and VfoldLA.Methods Mol Biol. 2021;2323:1-11. doi: 10.1007/978-1-0716-1499-0_1. Methods Mol Biol. 2021. PMID: 34086269 Free PMC article.
-
Vfold: a web server for RNA structure and folding thermodynamics prediction.PLoS One. 2014 Sep 12;9(9):e107504. doi: 10.1371/journal.pone.0107504. eCollection 2014. PLoS One. 2014. PMID: 25215508 Free PMC article.
-
Automated modeling of RNA 3D structure.Methods Mol Biol. 2014;1097:395-415. doi: 10.1007/978-1-62703-709-9_18. Methods Mol Biol. 2014. PMID: 24639169 Review.
-
RNA Structure: Advances and Assessment of 3D Structure Prediction.Annu Rev Biophys. 2017 May 22;46:483-503. doi: 10.1146/annurev-biophys-070816-034125. Epub 2017 Mar 30. Annu Rev Biophys. 2017. PMID: 28375730 Review.
Cited by
-
trRosettaRNA: automated prediction of RNA 3D structure with transformer network.Nat Commun. 2023 Nov 9;14(1):7266. doi: 10.1038/s41467-023-42528-4. Nat Commun. 2023. PMID: 37945552 Free PMC article.
-
RNA-TorsionBERT: leveraging language models for RNA 3D torsion angles prediction.Bioinformatics. 2024 Dec 26;41(1):btaf004. doi: 10.1093/bioinformatics/btaf004. Bioinformatics. 2024. PMID: 39775709 Free PMC article.
-
Comparative analysis of RNA 3D structure prediction methods: towards enhanced modeling of RNA-ligand interactions.Nucleic Acids Res. 2024 Jul 22;52(13):7465-7486. doi: 10.1093/nar/gkae541. Nucleic Acids Res. 2024. PMID: 38917327 Free PMC article.
-
RNAbpFlow: Base pair-augmented SE(3)-flow matching for conditional RNA 3D structure generation.bioRxiv [Preprint]. 2025 Jan 26:2025.01.24.634669. doi: 10.1101/2025.01.24.634669. bioRxiv. 2025. PMID: 39896539 Free PMC article. Preprint.
-
RNA 3D Structure Prediction: Progress and Perspective.Molecules. 2023 Jul 20;28(14):5532. doi: 10.3390/molecules28145532. Molecules. 2023. PMID: 37513407 Free PMC article. Review.
References
-
- Cragnolini T. et al. (2015) Coarse-grained HiRE-RNA model for ab initio RNA folding beyond simple molecules, including noncanonical and multiple base pairings. J. Chem. Theory Comput., 11, 3510–3522. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

