Enterococcus faecalis Antagonizes Pseudomonas aeruginosa Growth in Mixed-Species Interactions
- PMID: 35758750
- PMCID: PMC9295543
- DOI: 10.1128/jb.00615-21
Enterococcus faecalis Antagonizes Pseudomonas aeruginosa Growth in Mixed-Species Interactions
Abstract
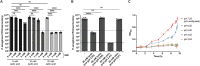
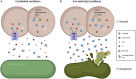
Enterococcus faecalis is often coisolated with Pseudomonas aeruginosa in polymicrobial biofilm-associated infections of wounds and the urinary tract. As a defense strategy, the host innately restricts iron availability at infection sites. Despite their coprevalence, the polymicrobial interactions of these two species in biofilms and under iron-restricted conditions remain unexplored. Here, we show that E. faecalis inhibits P. aeruginosa growth within biofilms when iron is restricted. E. faecalis lactate dehydrogenase (ldh1) gives rise to l-lactate production during fermentative growth. We find that an E. faecalis ldh1 mutant fails to inhibit P. aeruginosa growth. Additionally, we demonstrate that ldh1 expression is induced under iron-restricted conditions, resulting in increased lactic acid exported and, consequently, a reduction in local environmental pH. Together, our results suggest that E. faecalis synergistically inhibits P. aeruginosa growth by decreasing environmental pH and l-lactate-mediated iron chelation. Overall, this study emphasizes the importance of the microenvironment in polymicrobial interactions and how manipulating the microenvironment can impact the growth trajectory of bacterial communities. IMPORTANCE Many infections are polymicrobial and biofilm-associated in nature. Iron is essential for many metabolic processes and plays an important role in controlling infections, where the host restricts iron as a defense mechanism against invading pathogens. However, polymicrobial interactions between pathogens are underexplored under iron-restricted conditions. Here, we explore the polymicrobial interactions between commonly coisolated E. faecalis and P. aeruginosa within biofilms. We find that E. faecalis modulates the microenvironment by exporting lactic acid which further chelates already limited iron and also lowers the environmental pH to antagonize P. aeruginosa growth under iron-restricted conditions. Our findings provide insights into polymicrobial interactions between bacteria and how manipulating the microenvironment can be taken advantage of to better control infections.
Keywords: Enterococcus faecalis; Pseudomonas aeruginosa; iron restriction; l-lactate; lactate dehydrogenase (LDH); mixed species; polymicrobial interactions.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

