Identification of a Stable Chromosomal Tandem Multicopy of blaVIM-63, a New blaVIM-2 Carbapenemase
- PMID: 35758752
- PMCID: PMC9295573
- DOI: 10.1128/jb.00088-22
Identification of a Stable Chromosomal Tandem Multicopy of blaVIM-63, a New blaVIM-2 Carbapenemase
Abstract
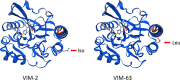
This study characterizes a new genetic structure containing a multicopy of a blaVIM-2 variant with an A676C substitution, blaVIM-63. This gene was detected on the chromosome of two carbapenem-resistant clinical strains of Citrobacter freundii ST22 recovered from two patients, separated by a 6-month period, and previously in Pseudomonas aeruginosa ST2242 from the same hospital unit. Short-read sequencing was used to characterize the new variant in both species, and long-read sequencing was used to characterize the genome of C. freundii. On the P. aeruginosa chromosome, the blaVIM-63 gene was inserted between ISPsy 42-type sequences, flanked by an intl1 sequence, nearby aph(3')-VI, and sul1. On the C. freundii chromosome, the blaVIM-63 gene was inserted into a Tn6230-like transposon as a stable five-tandem-repeat multimer, flanked by the same intl1 as in P. aeruginosa. This structure was stable across subcultures and did not change in the presence of carbapenems. The blaVIM-63 gene was cloned into the pCR-Blunt plasmid to study antimicrobial susceptibility patterns and into pET29a for kinetic activity analysis. VIM-63 showed higher Km values than VIM-2 for ceftazidime and cefepime and higher kcat values for cefotaxime, ceftazidime, imipenem, and ertapenem, without differences in MIC values. This is the first study to describe this new variant, VIM-63, in two different species with a chromosomal location integrated into different mobile elements and the first to describe a stable multimer of a metallo-β-lactamase. Despite the amino acid substitution, the susceptibility pattern of the new variant was similar to that of VIM-2. IMPORTANCE VIM group metallo-β-lactamases are usually captured by IntI1 integrases. This work describes the detection for the first time of a novel, previously unknown variant of VIM-2, VIM-63. This carbapenemase has been found on the chromosome of two different species, Citrobacter freundii and Pseudomonas aeruginosa, from the same hospital. The adjacent genetic environment of the blaVIM-63 gene would indicate that the capture of this gene by IntI1 has occurred in two different genetic events in each of the species, and in one there has been a stable integration of tandem copies of this gene.
Keywords: VIM-2 variant; VIM-63; WGS; carbapenemase; multimer.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

