Variants in Bedaquiline-Candidate-Resistance Genes: Prevalence in Bedaquiline-Naive Patients, Effect on MIC, and Association with Mycobacterium tuberculosis Lineage
- PMID: 35758754
- PMCID: PMC9295546
- DOI: 10.1128/aac.00322-22
Variants in Bedaquiline-Candidate-Resistance Genes: Prevalence in Bedaquiline-Naive Patients, Effect on MIC, and Association with Mycobacterium tuberculosis Lineage
Abstract
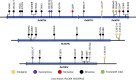
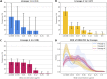
Studies have shown that variants in bedaquiline-resistance genes can occur in isolates from bedaquiline-naive patients. We assessed the prevalence of variants in all bedaquiline-candidate-resistance genes in bedaquiline-naive patients, investigated the association between these variants and lineage, and the effect on phenotype. We used whole-genome sequencing to identify variants in bedaquiline-resistance genes in isolates from 509 bedaquiline treatment naive South African tuberculosis patients. A phylogenetic tree was constructed to investigate the association with the isolate lineage background. Bedaquiline MIC was determined using the UKMYC6 microtiter assay. Variants were identified in 502 of 509 isolates (98.6%), with the highest (85%) prevalence of variants in the Rv0676c (mmpL5) gene. We identified 36 unique variants, including 19 variants not reported previously. Only four isolates had a bedaquiline MIC equal to or above the epidemiological cut-off value of 0.25 μg/mL. Phylogenetic analysis showed that 14 of the 15 variants observed more than once occurred monophyletically in one Mycobacterium tuberculosis (sub)lineage. The bedaquiline MIC differed between isolates belonging to lineage 2 and 4 (Fisher's exact test, P = 0.0004). The prevalence of variants in bedaquiline-resistance genes in isolates from bedaquiline-naive patients is high, but very few (<2%) isolates were phenotypically resistant. We found an association between variants in bedaquiline resistance genes and Mycobacterium tuberculosis (sub)lineage, resulting in a lineage-dependent difference in bedaquiline phenotype. Future studies should investigate the impact of the presence of variants on bedaquiline-resistance acquisition and treatment outcome.
Keywords: antibiotic resistance; antimicrobial resistance; bedaquiline; drug-resistant tuberculosis; epidemiology; phenotypic drug susceptibility testing; phylogeny; tuberculosis; whole-genome sequencing.
Conflict of interest statement
The authors declare a conflict of interest. K.L. reports funding by Janssen, Pharmaceutical Companies of Johnson & Johnson. T.C.R. reports funding by FIND (Geneva, Switzerland) and NIH/NIAD [grant number R21AI135756] and is co-inventor on a patent associated with the processing of sequencing files (European Patent Application No. 14840432.0 & USSN 14/912,918) and a provisional patent for a TB diagnostic assay (USSN Provisional Patent No. 63/048.989). R.M.W. reports baseline funding of the South African Medical Research Council. A.V.R. reports funding by Research Foundation Flanders [grant number G0F8316N].
Figures


References
-
- Zimenkov DV, Nosova EY, Kulagina EV, Antonova OV, Arslanbaeva LR, Isakova AI, Krylova LY, Peretokina IV, Makarova MV, Safonova SG, Borisov SE, Gryadunov DA. 2017. Examination of bedaquiline- and linezolid-resistant Mycobacterium tuberculosis isolates from the Moscow region. J Antimicrob Chemother 72:1901–1906. 10.1093/jac/dkx094. - DOI - PubMed
-
- Bloemberg GV, Keller PM, Stucki D, Stuckia D, Trauner A, Borrell S, Latshang T, Coscolla M, Rothe T, Hömke R, Ritter C, Feldmann J, Schulthess B, Gagneux S, Böttger EC. 2015. Acquired resistance to bedaquiline and delamanid in therapy for tuberculosis. N Engl J Med 373:1986–1988. 10.1056/NEJMc1505196. - DOI - PMC - PubMed
-
- World Health Organization. 2021. Catalogue of mutations in Mycobacterium tuberculosis complex and their association with drug resistance. WHO.

