The minimizer Jaccard estimator is biased and inconsistent
- PMID: 35758786
- PMCID: PMC9235516
- DOI: 10.1093/bioinformatics/btac244
The minimizer Jaccard estimator is biased and inconsistent
Abstract
Motivation: Sketching is now widely used in bioinformatics to reduce data size and increase data processing speed. Sketching approaches entice with improved scalability but also carry the danger of decreased accuracy and added bias. In this article, we investigate the minimizer sketch and its use to estimate the Jaccard similarity between two sequences.
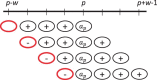
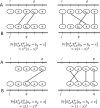
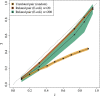
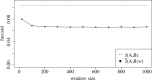
Results: We show that the minimizer Jaccard estimator is biased and inconsistent, which means that the expected difference (i.e. the bias) between the estimator and the true value is not zero, even in the limit as the lengths of the sequences grow. We derive an analytical formula for the bias as a function of how the shared k-mers are laid out along the sequences. We show both theoretically and empirically that there are families of sequences where the bias can be substantial (e.g. the true Jaccard can be more than double the estimate). Finally, we demonstrate that this bias affects the accuracy of the widely used mashmap read mapping tool.
Availability and implementation: Scripts to reproduce our experiments are available at https://github.com/medvedevgroup/minimizer-jaccard-estimator/tree/main/reproduce.
Supplementary information: Supplementary data are available at Bioinformatics online.
© The Author(s) 2022. Published by Oxford University Press.
Figures


References
-
- Blanca A. et al. (2021) The statistics of k-mers from a sequence undergoing a simple mutation process without spurious matches. J. Comput. Biol., 29, 155–168. https://www.liebertpub.com/doi/10.1089/cmb.2021.0431. - DOI - PMC - PubMed
-
- Broder A.Z. (1997) On the resemblance and containment of documents. In: Proceedings. Compression and Complexity of SEQUENCES 1997 (Cat. No. 97TB100171). IEEE, pp. 21–29.
-
- Chin C.-S., Khalak A. (2019) Human genome assembly in 100 minutes. bioRxiv, page 705616.
-
- Cormode G., Muthukrishnan S. (2004) An improved data stream summary: the count-min sketch and its applications. In: Latin American Symposium on Theoretical Informatics. Springer, pp. 29–38.
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources

