CALDERA: finding all significant de Bruijn subgraphs for bacterial GWAS
- PMID: 35758804
- PMCID: PMC9235473
- DOI: 10.1093/bioinformatics/btac238
CALDERA: finding all significant de Bruijn subgraphs for bacterial GWAS
Abstract
Motivation: Genome-wide association studies (GWAS), aiming to find genetic variants associated with a trait, have widely been used on bacteria to identify genetic determinants of drug resistance or hypervirulence. Recent bacterial GWAS methods usually rely on k-mers, whose presence in a genome can denote variants ranging from single-nucleotide polymorphisms to mobile genetic elements. This approach does not require a reference genome, making it easier to account for accessory genes. However, a same gene can exist in slightly different versions across different strains, leading to diluted effects.
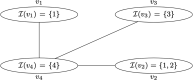
Results: Here, we overcome this issue by testing covariates built from closed connected subgraphs (CCSs) of the de Bruijn graph defined over genomic k-mers. These covariates capture polymorphic genes as a single entity, improving k-mer-based GWAS both in terms of power and interpretability. However, a method naively testing all possible subgraphs would be powerless due to multiple testing corrections, and the mere exploration of these subgraphs would quickly become computationally intractable. The concept of testable hypothesis has successfully been used to address both problems in similar contexts. We leverage this concept to test all CCSs by proposing a novel enumeration scheme for these objects which fully exploits the pruning opportunity offered by testability, resulting in drastic improvements in computational efficiency. Our method integrates with existing visual tools to facilitate interpretation.
Availability and implementation: We provide an implementation of our method, as well as code to reproduce all results at https://github.com/HectorRDB/Caldera_ISMB.
Supplementary information: Supplementary data are available at Bioinformatics online.
© The Author(s) 2022. Published by Oxford University Press.
Figures

References
-
- Avis D., Fukuda K. (1996) Reverse search for enumeration. Discrete Appl. Math., 65, 21–46.
-
- Bonferroni C. (1936) Teoria Statistica Delle Classi e Calcolo Delle Probabilità. Pubblicazioni del R Istituto Superiore di Scienze Economiche e Commerciali di Firenze, Vol. 8. Seeber, pp. 3–62.
-
- de Bruijn N. (1946) A combinatorial problem. Proc. Sect. Sci. K. Ned. Akad. Wet. Amst., 49, 758–764.

