How Microtubules Build the Spindle Branch by Branch
- PMID: 35759800
- PMCID: PMC9619725
- DOI: 10.1146/annurev-cellbio-120420-114559
How Microtubules Build the Spindle Branch by Branch
Abstract
The microtubule (MT) cytoskeleton provides the architecture that governs intracellular organization and the regulated motion of macromolecules through the crowded cytoplasm. The key to establishing a functioning cytoskeletal architecture is regulating when and where new MTs are nucleated. Within the spindle, the vast majority of MTs are generated through a pathway known as branching MT nucleation, which exponentially amplifies MT number in a polar manner. Whereas other MT nucleation pathways generally require a complex organelle such as the centrosome or Golgi apparatus to localize nucleation factors, the branching site is based solely on a simple, preformed MT, making it an ideal system to study MT nucleation. In this review, we address recent developments in characterizing branching factors, the branching reaction, and its regulation, as well as branching MT nucleation in systems beyond the spindle and within human disease.
Keywords: TPX2; augmin; branching microtubule nucleation; meiosis; mitosis; γ-TuRC; γ-tubulin ring complex.
Figures


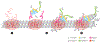

References
-
- Boissan M, Schlattner U, Lacombe ML. 2018. The NDPK/NME superfamily: state of the art. Lab. Investig 98:164–74 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

