Discovery of 2-thiobenzimidazoles as noncovalent inhibitors of SARS-CoV-2 main protease
- PMID: 35760254
- PMCID: PMC9225965
- DOI: 10.1016/j.bmcl.2022.128867
Discovery of 2-thiobenzimidazoles as noncovalent inhibitors of SARS-CoV-2 main protease
Abstract
The discovery of antiviral agents against SARS-CoV-2 is an important step toward ending the COVID-19 pandemic and to tackle future outbreaks. In this context, the main protease (Mpro) represents an ideal target for developing coronavirus antivirals, being conserved among different strains and essential for survival. In this work, using in silico tools, we created and validated a docking protocol able to predict binders to the catalytic site of Mpro. The following structure-based virtual screening of a subset of the ZINC library (over 4.3 million unique structures), led to the identification of a hit compound having a 2-thiobenzimidazole scaffold. The inhibitory activity was confirmed using a FRET-based proteolytic assay against recombinant Mpro. Structure-activity relationships were obtained with the synthesis of a small library of analogs, guided by the analysis of the docking pose. Our efforts led to the identification of a micromolar Mpro inhibitor (IC50 = 14.9 µM) with an original scaffold possessing ideal drug-like properties (predicted using the QikProp function) and representing a promising lead for the development of a novel class of coronavirus antivirals.
Copyright © 2022 Elsevier Ltd. All rights reserved.
Conflict of interest statement
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Figures

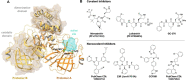
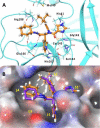
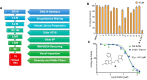

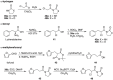
References
-
- World Health Organization. WHO Coronavirus (COVID-19) Dashboard. https://covid19.who.int/ (accessed April 26, 2022).
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Chemical Information
Miscellaneous

