Multiple epigenetic factors co-localize with HMGN proteins in A-compartment chromatin
- PMID: 35761366
- PMCID: PMC9235084
- DOI: 10.1186/s13072-022-00457-4
Multiple epigenetic factors co-localize with HMGN proteins in A-compartment chromatin
Abstract
Background: Nucleosomal binding proteins, HMGN, is a family of chromatin architectural proteins that are expressed in all vertebrate nuclei. Although previous studies have discovered that HMGN proteins have important roles in gene regulation and chromatin accessibility, whether and how HMGN proteins affect higher order chromatin status remains unknown.
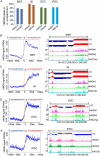
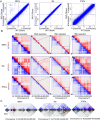
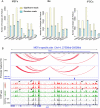
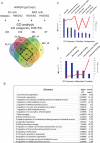
Results: We examined the roles that HMGN1 and HMGN2 proteins play in higher order chromatin structures in three different cell types. We interrogated data generated in situ, using several techniques, including Hi-C, Promoter Capture Hi-C, ChIP-seq, and ChIP-MS. Our results show that HMGN proteins occupy the A compartment in the 3D nucleus space. In particular, HMGN proteins occupy genomic regions involved in cell-type-specific long-range promoter-enhancer interactions. Interestingly, depletion of HMGN proteins in the three different cell types does not cause structural changes in higher order chromatin, i.e., in topologically associated domains (TADs) and in A/B compartment scores. Using ChIP-seq combined with mass spectrometry, we discovered protein partners that are directly associated with or neighbors of HMGNs on nucleosomes.
Conclusions: We determined how HMGN chromatin architectural proteins are positioned within a 3D nucleus space, including the identification of their binding partners in mononucleosomes. Our research indicates that HMGN proteins localize to active chromatin compartments but do not have major effects on 3D higher order chromatin structure and that their binding to chromatin is not dependent on specific protein partners.
Keywords: Chromatin structure; HMGN; Hi–C; Mass spectrometry.
© 2022. This is a U.S. Government work and not under copyright protection in the US; foreign copyright protection may apply.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures

References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

