Genetic prediction of impulse control disorders in Parkinson's disease
- PMID: 35762106
- PMCID: PMC9268896
- DOI: 10.1002/acn3.51569
Genetic prediction of impulse control disorders in Parkinson's disease
Abstract
Objective: To develop a clinico-genetic predictor of impulse control disorder (ICD) risk in Parkinson's disease (PD).
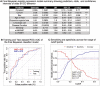
Methods: In 5770 individuals from three PD cohorts (the 23andMe, Inc.; the University of Pennsylvania [UPenn]; and the Parkinson's Progression Markers Initiative [PPMI]), we used a discovery-replication strategy to develop a clinico-genetic predictor for ICD risk. We first performed a Genomewide Association Study (GWAS) for ICDs anytime during PD in 5262 PD individuals from the 23andMe cohort. We then combined newly discovered ICD risk loci with 13 ICD risk loci previously reported in the literature to develop a model predicting ICD in a Training dataset (n = 339, from UPenn and PPMI cohorts). The model was tested in a non-overlapping Test dataset (n = 169, from UPenn and PPMI cohorts) and used to derive a continuous measure, the ICD-risk score (ICD-RS), enriching for PD individuals with ICD (ICD+ PD).
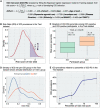
Results: By GWAS, we discovered four new loci associated with ICD at p-values of 4.9e-07 to 1.3e-06. Our best logistic regression model included seven clinical and two genetic variables, achieving an area under the receiver operating curve for ICD prediction of 0.75 in the Training and 0.72 in the Test dataset. The ICD-RS separated groups of PD individuals with ICD prevalence of nearly 40% (highest risk quartile) versus 7% (lowest risk quartile).
Interpretation: In this multi-cohort, international study, we developed an easily computed clinico-genetic tool, the ICD-RS, that substantially enriches for subgroups of PD at very high versus very low risk for ICD, enabling pharmacogenetic approaches to PD medication selection.
© 2022 The Authors. Annals of Clinical and Translational Neurology published by Wiley Periodicals LLC on behalf of American Neurological Association.
Conflict of interest statement
Alice S. Chen‐Plotkin, Marijan Posavi, and Daniel Weintraub declare that they are the inventors of a University of Pennsylvania patent (pending) covering prediction of impulsivity in Parkinson's Disease.
Pierre Fontanillas and Paul Cannon are employed by and hold stock or stock options in 23andMe, Inc.
Figures



References
-
- Voon V, Fox SH. Medication‐related impulse control and repetitive behaviors in Parkinson disease. Arch Neurol. 2007;64(8):1089‐1096. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

