Exploring antiviral and anti-inflammatory effects of thiol drugs in COVID-19
- PMID: 35762590
- PMCID: PMC9448286
- DOI: 10.1152/ajplung.00136.2022
Exploring antiviral and anti-inflammatory effects of thiol drugs in COVID-19
Abstract
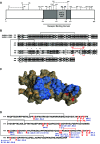
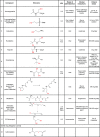
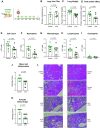
The redox status of the cysteine-rich SARS-CoV-2 spike glycoprotein (SARS-2-S) is important for the binding of SARS-2-S to angiotensin-converting enzyme 2 (ACE2), suggesting that drugs with a functional thiol group ("thiol drugs") may cleave cystines to disrupt SARS-CoV-2 cell entry. In addition, neutrophil-induced oxidative stress is a mechanism of COVID-19 lung injury, and the antioxidant and anti-inflammatory properties of thiol drugs, especially cysteamine, may limit this injury. To first explore the antiviral effects of thiol drugs in COVID-19, we used an ACE-2 binding assay and cell entry assays utilizing reporter pseudoviruses and authentic SARS-CoV-2 viruses. We found that multiple thiol drugs inhibit SARS-2-S binding to ACE2 and virus infection. The most potent drugs were effective in the low millimolar range, and IC50 values followed the order of their cystine cleavage rates and lower thiol pKa values. To determine if thiol drugs have antiviral effects in vivo and to explore any anti-inflammatory effects of thiol drugs in COVID-19, we tested the effects of cysteamine delivered intraperitoneally to hamsters infected with SARS-CoV-2. Cysteamine did not decrease lung viral infection, but it significantly decreased lung neutrophilic inflammation and alveolar hemorrhage. We speculate that the concentration of cysteamine achieved in the lungs with intraperitoneal delivery was insufficient for antiviral effects but sufficient for anti-inflammatory effects. We conclude that thiol drugs decrease SARS-CoV-2 lung inflammation and injury, and we provide rationale for future studies to test if direct (aerosol) delivery of thiol drugs to the airways might also result in antiviral effects.
Keywords: ACE2; COVID-19; SARS-CoV-2; cysteamine; thiol drugs.
Conflict of interest statement
J. V. Fahy, S. Oscarson, I. Gitlin, and W. W. Raymond are inventors on patent applications related to the use of thiol drugs as treatments for COVID-19. None of the other authors has any conflicts of interest, financial or otherwise, to disclose.
Figures




References
-
- Zhu N, Zhang D, Wang W, Li X, Yang B, Song J, Zhao X, Huang B, Shi W, Lu R, Niu P, Zhan F, Ma X, Wang D, Xu W, Wu G, Gao GF, Tan W; China Novel Coronavirus Investigating and Research Team. A novel coronavirus from patients with pneumonia in China, 2019. N Engl J Med 382: 727–733, 2020. doi:10.1056/NEJMoa2001017. - DOI - PMC - PubMed
-
- Hoffmann M, Kleine-Weber H, Schroeder S, Krüger N, Herrler T, Erichsen S, Schiergens TS, Herrler G, Wu NH, Nitsche A, Müller MA, Drosten C, Pöhlmann S. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell 181: 271–280.e8, 2020. doi:10.1016/j.cell.2020.02.052. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Associated data
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

