Segmentation and Evaluation of Corneal Nerves and Dendritic Cells From In Vivo Confocal Microscopy Images Using Deep Learning
- PMID: 35762938
- PMCID: PMC9251793
- DOI: 10.1167/tvst.11.6.24
Segmentation and Evaluation of Corneal Nerves and Dendritic Cells From In Vivo Confocal Microscopy Images Using Deep Learning
Abstract
Purpose: Segmentation and evaluation of in vivo confocal microscopy (IVCM) images requires manual intervention, which is time consuming, laborious, and non-reproducible. The aim of this research was to develop and validate deep learning-based methods that could automatically segment and evaluate corneal nerve fibers (CNFs) and dendritic cells (DCs) in IVCM images, thereby reducing processing time to analyze larger volumes of clinical images.
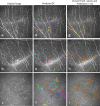
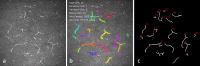
Methods: CNF and DC segmentation models were developed based on U-Net and Mask R-CNN architectures, respectively; 10-fold cross-validation was used to evaluate both models. The CNF model was trained and tested using 1097 and 122 images, and the DC model was trained and tested using 679 and 75 images, respectively, at each fold. The CNF morphology, number of nerves, number of branching points, nerve length, and tortuosity were analyzed; for DCs, number, size, and immature-mature cells were analyzed. Python-based software was written for model training, testing, and automatic morphometric parameters evaluation.
Results: The CNF model achieved on average 86.1% sensitivity and 90.1% specificity, and the DC model achieved on average 89.37% precision, 94.43% recall, and 91.83% F1 score. The interclass correlation coefficient (ICC) between manual annotation and automatic segmentation were 0.85, 0.87, 0.95, and 0.88 for CNF number, length, branching points, and tortuosity, respectively, and the ICC for DC number and size were 0.95 and 0.92, respectively.
Conclusions: Our proposed methods demonstrated reliable consistency between manual annotation and automatic segmentation of CNF and DC with rapid speed. The results showed that these approaches have the potential to be implemented into clinical practice in IVCM images.
Translational relevance: The deep learning-based automatic segmentation and quantification algorithm significantly increases the efficiency of evaluating IVCM images, thereby supporting and potentially improving the diagnosis and treatment of ocular surface disease associated with corneal nerves and dendritic cells.
Conflict of interest statement
Disclosure:
Figures








References
-
- Craig JP, Nelson JD, Azar DT, et al.. TFOS DEWS II Report Executive Summary. Ocul Surf. 2017; 15(4): 802–812. - PubMed
-
- Stapleton F, Alves M, Bunya VY, et al.. TFOS DEWS II Epidemiology Report. Ocul Surf. 2017; 15(3): 334–365. - PubMed
-
- Craig JP, Nichols KK, Akpek EK, et al.. TFOS DEWS II Definition and Classification Report. Ocul Surf. 2017; 15(3): 276–283. - PubMed
-
- McDonald M, Patel DA, Keith MS, Snedecor SJ.. Economic and humanistic burden of dry eye disease in Europe, North America, and Asia: a systematic literature review. Ocul Surf. 2016; 14(2): 144–167. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources

