The RNA-binding protein Swm is critical for Drosophila melanogaster intestinal progenitor cell maintenance
- PMID: 35762963
- PMCID: PMC9526066
- DOI: 10.1093/genetics/iyac099
The RNA-binding protein Swm is critical for Drosophila melanogaster intestinal progenitor cell maintenance
Abstract
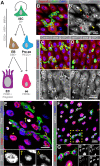
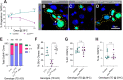
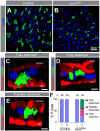
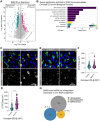
The regulation of stem cell survival, self-renewal, and differentiation is critical for the maintenance of tissue homeostasis. Although the involvement of signaling pathways and transcriptional control mechanisms in stem cell regulation have been extensively investigated, the role of post-transcriptional control is still poorly understood. Here, we show that the nuclear activity of the RNA-binding protein Second Mitotic Wave Missing is critical for Drosophila melanogaster intestinal stem cells and their daughter cells, enteroblasts, to maintain their progenitor cell properties and functions. Loss of swm causes intestinal stem cells and enteroblasts to stop dividing and instead detach from the basement membrane, resulting in severe progenitor cell loss. swm loss is further characterized by nuclear accumulation of poly(A)+ RNA in progenitor cells. Second Mitotic Wave Missing associates with transcripts involved in epithelial cell maintenance and adhesion, and the loss of swm, while not generally affecting the levels of these Second Mitotic Wave Missing-bound mRNAs, leads to elevated expression of proteins encoded by some of them, including the fly ortholog of Filamin. Taken together, this study indicates a nuclear role for Second Mitotic Wave Missing in adult stem cell maintenance, raising the possibility that nuclear post-transcriptional regulation of mRNAs encoding cell adhesion proteins ensures proper attachment of progenitor cells.
Keywords: cheerio; rhea; swm; cell adhesion; intestinal stem cells.
© The Author(s) 2022. Published by Oxford University Press on behalf of Genetics Society of America. All rights reserved. For permissions, please email: journals.permissions@oup.com.
Figures

References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

