Outlier detection for multi-network data
- PMID: 35762974
- PMCID: PMC9890313
- DOI: 10.1093/bioinformatics/btac431
Outlier detection for multi-network data
Abstract
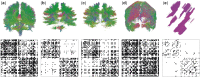
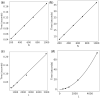
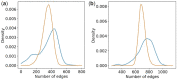
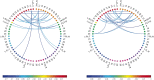
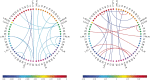
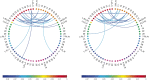
Motivation: It has become routine in neuroscience studies to measure brain networks for different individuals using neuroimaging. These networks are typically expressed as adjacency matrices, with each cell containing a summary of connectivity between a pair of brain regions. There is an emerging statistical literature describing methods for the analysis of such multi-network data in which nodes are common across networks but the edges vary. However, there has been essentially no consideration of the important problem of outlier detection. In particular, for certain subjects, the neuroimaging data are so poor quality that the network cannot be reliably reconstructed. For such subjects, the resulting adjacency matrix may be mostly zero or exhibit a bizarre pattern not consistent with a functioning brain. These outlying networks may serve as influential points, contaminating subsequent statistical analyses. We propose a simple Outlier DetectIon for Networks (ODIN) method relying on an influence measure under a hierarchical generalized linear model for the adjacency matrices. An efficient computational algorithm is described, and ODIN is illustrated through simulations and an application to data from the UK Biobank.
Results: ODIN was successful in identifying moderate to extreme outliers. Removing such outliers can significantly change inferences in downstream applications.
Availability and implementation: ODIN has been implemented in both Python and R and these implementations along with other code are publicly available at github.com/pritamdey/ODIN-python and github.com/pritamdey/ODIN-r, respectively.
Supplementary information: Supplementary data are available at Bioinformatics online.
© The Author(s) 2022. Published by Oxford University Press. All rights reserved. For permissions, please e-mail: journals.permissions@oup.com.
Figures

References
-
- Aliverti E., Durante D. (2019) Spatial modeling of brain connectivity data via latent distance models with nodes clustering. Stat. Anal. Data Min. ASA Data Sci. J., 12, 185–196.
-
- Desikan R.S. et al. (2006) An automated labeling system for subdividing the human cerebral cortex on MRI scans into gyral based regions of interest. NeuroImage, 31, 968–980. - PubMed

