Structural basis of Omicron immune evasion: A comparative computational study
- PMID: 35763933
- PMCID: PMC9212419
- DOI: 10.1016/j.compbiomed.2022.105758
Structural basis of Omicron immune evasion: A comparative computational study
Abstract
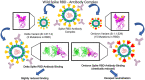
Background: The vaccines used against SARS-CoV-2 by now have been able to develop some neutralising antibodies in the vaccinated population and their effectiveness has been challenged by the emergence of the new strains with numerous mutations in the spike protein of SARS-CoV-2. Since S protein is the major immunogenic protein of the virus which contains Receptor Binding Domain (RBD) that interacts with the human Angiotensin-Converting Enzyme 2 (ACE2) receptors, any mutations in this region should affect the neutralisation potential of the antibodies leading to the immune evasion. Several variants of concern of the virus have emerged so far, amongst which the most critical are Delta and recently reported Omicron. In this study, we have mapped and reported mutations on the modelled RBD and evaluated binding affinities of various human antibodies with it.
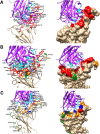
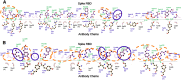
Method: Docking and molecular dynamics simulation studies have been used to explore the effect of mutations on the structure of RBD and RBD-antibody interaction.
Results: These analyses show that the mutations mostly at the interface of a nearby region lower the binding affinity of the antibody by ten to forty percent, with a downfall in the number of interactions formed as a whole. It implies the generation of immune escape variants.
Conclusions: Notable mutations and their effect was characterised that explain the structural basis of antibody efficacy in Delta and a compromised neutralisation effect for the Omicron variant. Thus, our results pave the way for robust vaccine design that can be effective for many variants.
Keywords: Compromised neutralisation; Molecular docking; Molecular dynamics simulations; Omicron; SARS-CoV-2 variants.
Copyright © 2022 Elsevier Ltd. All rights reserved.
Conflict of interest statement
Authors declare no conflict of interest.
Figures




References
-
- Cascella M., Rajnik M., Aleem A., Dulebohn S.C., Di Napoli R. Treasure Island. 2022. Features, evaluation, and treatment of coronavirus (COVID-19) (FL) - PubMed
Publication types
MeSH terms
Substances
Supplementary concepts
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

