Multimodal data integration using machine learning improves risk stratification of high-grade serous ovarian cancer
- PMID: 35764743
- PMCID: PMC9239907
- DOI: 10.1038/s43018-022-00388-9
Multimodal data integration using machine learning improves risk stratification of high-grade serous ovarian cancer
Abstract
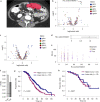
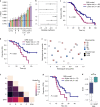
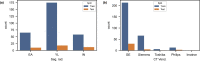
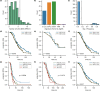
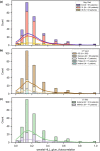
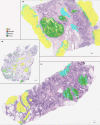
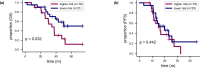
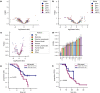
Patients with high-grade serous ovarian cancer suffer poor prognosis and variable response to treatment. Known prognostic factors for this disease include homologous recombination deficiency status, age, pathological stage and residual disease status after debulking surgery. Recent work has highlighted important prognostic information captured in computed tomography and histopathological specimens, which can be exploited through machine learning. However, little is known about the capacity of combining features from these disparate sources to improve prediction of treatment response. Here, we assembled a multimodal dataset of 444 patients with primarily late-stage high-grade serous ovarian cancer and discovered quantitative features, such as tumor nuclear size on staining with hematoxylin and eosin and omental texture on contrast-enhanced computed tomography, associated with prognosis. We found that these features contributed complementary prognostic information relative to one another and clinicogenomic features. By fusing histopathological, radiologic and clinicogenomic machine-learning models, we demonstrate a promising path toward improved risk stratification of patients with cancer through multimodal data integration.
© 2022. The Author(s).
Conflict of interest statement
S.P.S. is a shareholder and consultant to Imagia Canexia Health Inc. Y.L is a shareholder of Y-mAbs Therapeutics Inc. and a consultant to Calyx. J.S.R.-F. reports receiving personal/consultancy fees from Goldman Sachs, REPARE Therapeutics, Paige.AI and Eli Lilly, membership of the scientific advisory boards of VolitionRx, REPARE Therapeutics and Paige.AI, membership of the Board of Directors of Grupo Oncoclinicas and ad hoc membership of the scientific advisory boards of Roche Tissue Diagnostics, Ventana Medical Systems, Novartis, Genentech and InVicro. J.S.R.-F. owns Paige.AI stock options. The other authors declare no competing interests.
Figures







Comment in
-
Ovarian cancer through a multi-modal lens.Nat Cancer. 2022 Jun;3(6):662-664. doi: 10.1038/s43018-022-00397-8. Nat Cancer. 2022. PMID: 35764744 No abstract available.
References
-
- National Cancer Institute. Cancer Stat Facts. https://seer.cancer.gov/statfacts/
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

