Landmark-based retrieval of inflamed skin vessels enabled by 3D correlative intravital light and volume electron microscopy
- PMID: 35764846
- PMCID: PMC9338004
- DOI: 10.1007/s00418-022-02119-8
Landmark-based retrieval of inflamed skin vessels enabled by 3D correlative intravital light and volume electron microscopy
Abstract
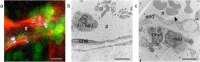
The nanometer spatial resolution of electron microscopy imaging remains an advantage over light microscopy, but the restricted field of view that can be inspected and the inability to visualize dynamic cellular events are definitely drawbacks of standard transmission electron microscopy (TEM). Several methods have been developed to overcome these limitations, mainly by correlating the light microscopical image to the electron microscope with correlative light and electron microscopy (CLEM) techniques. Since there is more than one method to obtain the region of interest (ROI), the workflow must be adjusted according to the research question and biological material addressed. Here, we describe in detail the development of a three-dimensional CLEM workflow for mouse skin tissue exposed to an inflammation stimulus and imaged by intravital microscopy (IVM) before fixation. Our aim is to relocate a distinct vessel in the electron microscope, addressing a complex biological question: how do cells interact with each other and the surrounding environment at the ultrastructural level? Retracing the area over several preparation steps did not involve any specific automated instruments but was entirely led by anatomical and artificially introduced landmarks, including blood vessel architecture and carbon-coated grids. Successful retrieval of the ROI by electron microscopy depended on particularly high precision during sample manipulation and extensive documentation. Further modification of the TEM sample preparation protocol for mouse skin tissue even rendered the specimen suitable for serial block-face scanning electron microscopy (SBF-SEM).
Keywords: Correlative light and electron microscopy; Dorsal skinfold chamber; Intravital microscopy; Live cell imaging; Serial block-face scanning electron microscopy; Transmission electron microscopy.
© 2022. The Author(s).
Conflict of interest statement
The authors declare no competing interest.
Figures



Similar articles
-
A workflow for 3D-CLEM investigating liver tissue.J Microsc. 2021 Mar;281(3):231-242. doi: 10.1111/jmi.12967. Epub 2020 Oct 27. J Microsc. 2021. PMID: 33034376
-
Reactive oxygen FIB spin milling enables correlative workflow for 3D super-resolution light microscopy and serial FIB/SEM of cultured cells.Sci Rep. 2021 Jun 23;11(1):13162. doi: 10.1038/s41598-021-92608-y. Sci Rep. 2021. PMID: 34162977 Free PMC article.
-
Correlative two-photon and serial block face scanning electron microscopy in neuronal tissue using 3D near-infrared branding maps.Methods Cell Biol. 2017;140:245-276. doi: 10.1016/bs.mcb.2017.03.007. Epub 2017 Apr 19. Methods Cell Biol. 2017. PMID: 28528636
-
Correlative light and volume electron microscopy to study brain development.Microscopy (Oxf). 2023 Aug 4;72(4):279-286. doi: 10.1093/jmicro/dfad002. Microscopy (Oxf). 2023. PMID: 36620906 Review.
-
Correlation of organelle dynamics between light microscopic live imaging and electron microscopic 3D architecture using FIB-SEM.Microscopy (Oxf). 2021 Mar 24;70(2):161-170. doi: 10.1093/jmicro/dfaa071. Microscopy (Oxf). 2021. PMID: 33216938 Free PMC article. Review.
Cited by
-
The extracellular matrix dictates regional competence for tumour initiation.Nature. 2023 Nov;623(7988):828-835. doi: 10.1038/s41586-023-06740-y. Epub 2023 Nov 15. Nature. 2023. PMID: 37968399 Free PMC article.
-
In focus in HCB.Histochem Cell Biol. 2022 Aug;158(2):123-125. doi: 10.1007/s00418-022-02142-9. Histochem Cell Biol. 2022. PMID: 35859014 No abstract available.
References
-
- Currie SM, Stegmeyer RI, Mildner K, Breitsprecher L, Zeuschner D, Psathaki OE, Schafer K, Wilkens M, Volkery S, Nieswandt B, Vestweber D. Confocal real-time analysis of cutaneous platelet recruitment during immune-complex-mediated inflammation. J Invest Dermatol. 2022 doi: 10.1016/j.jid.2022.03.011. - DOI - PubMed
-
- Deerinck TJ, Bushong EA, Thor A, Ellisman MH (2010) NCMIR methods for 3D EM: a new protocol for preparation of biological specimens for serial blockface scanning electron microscopy. https://ncmir.ucsd.edu/sbemprotocol
MeSH terms
LinkOut - more resources
Full Text Sources

