Development and validation of a trans-ancestry polygenic risk score for type 2 diabetes in diverse populations
- PMID: 35765100
- PMCID: PMC9241245
- DOI: 10.1186/s13073-022-01074-2
Development and validation of a trans-ancestry polygenic risk score for type 2 diabetes in diverse populations
Abstract
Background: Type 2 diabetes (T2D) is a worldwide scourge caused by both genetic and environmental risk factors that disproportionately afflicts communities of color. Leveraging existing large-scale genome-wide association studies (GWAS), polygenic risk scores (PRS) have shown promise to complement established clinical risk factors and intervention paradigms, and improve early diagnosis and prevention of T2D. However, to date, T2D PRS have been most widely developed and validated in individuals of European descent. Comprehensive assessment of T2D PRS in non-European populations is critical for equitable deployment of PRS to clinical practice that benefits global populations.
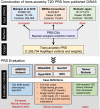
Methods: We integrated T2D GWAS in European, African, and East Asian populations to construct a trans-ancestry T2D PRS using a newly developed Bayesian polygenic modeling method, and assessed the prediction accuracy of the PRS in the multi-ethnic Electronic Medical Records and Genomics (eMERGE) study (11,945 cases; 57,694 controls), four Black cohorts (5137 cases; 9657 controls), and the Taiwan Biobank (4570 cases; 84,996 controls). We additionally evaluated a post hoc ancestry adjustment method that can express the polygenic risk on the same scale across ancestrally diverse individuals and facilitate the clinical implementation of the PRS in prospective cohorts.
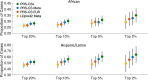
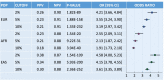
Results: The trans-ancestry PRS was significantly associated with T2D status across the ancestral groups examined. The top 2% of the PRS distribution can identify individuals with an approximately 2.5-4.5-fold of increase in T2D risk, which corresponds to the increased risk of T2D for first-degree relatives. The post hoc ancestry adjustment method eliminated major distributional differences in the PRS across ancestries without compromising its predictive performance.
Conclusions: By integrating T2D GWAS from multiple populations, we developed and validated a trans-ancestry PRS, and demonstrated its potential as a meaningful index of risk among diverse patients in clinical settings. Our efforts represent the first step towards the implementation of the T2D PRS into routine healthcare.
Keywords: Clinical implementation; Diverse populations; Polygenic risk score; Type 2 diabetes.
© 2022. The Author(s).
Conflict of interest statement
CYC is an employee of Biogen, Inc. and holds Biogen stock. KWC is involved in the Genetics and Neuroscience Special Interest Group and the Anxiety & Depression Association of America. JJC serves on the Board of Directors of the American Medical Informatics Association. EEK is a paid consultant of Encompass Bio and Galatea Bio, received honoraria from Illumina, Regeneron and 23&Me, and is a member of the Board of Directors of the American Society of Human Genetics and a member of the International Expert Committee, H3Africa. KK serves on the Scientific Advisory Board for Gilead Sciences and Goldfinch Bio. STW has received compensation from UpToDate. JWS is a member of the Leon Levy Foundation Neuroscience Advisory Board, participates on the Data and Safety Monitoring Board (DSMB) for the Implementing Genomics in Practice (IGNITE II) program, and received an honorarium for an internal seminar at Biogen, Inc. JBM is an Academic Associate with Quest Diagnostics R&D. EP is a paid consultant for Allelica. The remaining authors declare that they have no competing interests.
Figures
References
-
- Menke A, Casagrande S, Geiss L, Cowie CC. Prevalence of and trends in diabetes among adults in the United States, 1988-2012. JAMA. 2015;314(10):1021–1029. - PubMed
-
- Centers for Disease Control and Prevention . National Diabetes Statistics Report. 2020.
Publication types
MeSH terms
Grants and funding
- R00 AG054573/AG/NIA NIH HHS/United States
- U01 HG011167/HG/NHGRI NIH HHS/United States
- R03 DK131249/DK/NIDDK NIH HHS/United States
- R01 HL136666/HL/NHLBI NIH HHS/United States
- K24 HL133373/HL/NHLBI NIH HHS/United States
- R35 GM140487/GM/NIGMS NIH HHS/United States
- U01 HG011723/HG/NHGRI NIH HHS/United States
- U01 DK078616/DK/NIDDK NIH HHS/United States
- R35 HL155466/HL/NHLBI NIH HHS/United States
- U01HG008685/HG/NHGRI NIH HHS/United States
- U01 HG006379/HG/NHGRI NIH HHS/United States
- U01 HG006375/HG/NHGRI NIH HHS/United States
- U01 HG006830/HG/NHGRI NIH HHS/United States
- U01 NS041588/NS/NINDS NIH HHS/United States
- U01 HG011172/HG/NHGRI NIH HHS/United States
- U01 HG006388/HG/NHGRI NIH HHS/United States
- K23 DK114551/DK/NIDDK NIH HHS/United States
- R01 HL123782/HL/NHLBI NIH HHS/United States
- K25 DK128563/DK/NIDDK NIH HHS/United States
- UM1 DK078616/DK/NIDDK NIH HHS/United States
- U01 HG008685/HG/NHGRI NIH HHS/United States
- F32 HD108873/HD/NICHD NIH HHS/United States
LinkOut - more resources
Full Text Sources
Medical

