Meta-Analysis of Altered Gut Microbiota Reveals Microbial and Metabolic Biomarkers for Colorectal Cancer
- PMID: 35766483
- PMCID: PMC9431300
- DOI: 10.1128/spectrum.00013-22
Meta-Analysis of Altered Gut Microbiota Reveals Microbial and Metabolic Biomarkers for Colorectal Cancer
Abstract
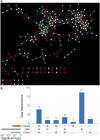
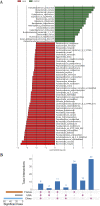
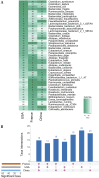
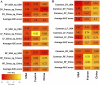
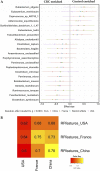
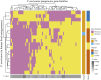
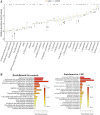
Colorectal cancer (CRC) is the second leading cause of cancer mortality worldwide. The dysbiotic gut microbiota and its metabolite secretions play a significant role in CRC development and progression. In this study, we identified microbial and metabolic biomarkers applicable to CRC using a meta-analysis of metagenomic datasets from diverse geographical regions. We used LEfSe, random forest (RF), and co-occurrence network methods to identify microbial biomarkers. Geographic dataset-specific markers were identified and evaluated using area under the ROC curve (AUC) scores and random effect size. Co-occurrence networks analysis showed a reduction in the overall microbial associations and the presence of oral pathogenic microbial clusters in CRC networks. Analysis of predicted metabolites from CRC datasets showed the enrichment of amino acids, cadaverine, and creatine in CRC, which were positively correlated with CRC-associated microbes (Peptostreptococcus stomatis, Gemella morbillorum, Bacteroides fragilis, Parvimonas spp., Fusobacterium nucleatum, Solobacterium moorei, and Clostridium symbiosum), and negatively correlated with control-associated microbes. Conversely, butyrate, nicotinamide, choline, tryptophan, and 2-hydroxybutanoic acid showed positive correlations with control-associated microbes (P < 0.05). Overall, our study identified a set of global CRC biomarkers that are reproducible across geographic regions. We also reported significant differential metabolites and microbe-metabolite interactions associated with CRC. This study provided significant insights for further investigations leading to the development of noninvasive CRC diagnostic tools and therapeutic interventions. IMPORTANCE Several studies showed associations between gut dysbiosis and CRC. Yet, the results are not conclusive due to cohort-specific associations that are influenced by genomic, dietary, and environmental stimuli and associated reproducibility issues with various analysis approaches. Emerging evidence suggests the role of microbial metabolites in modulating host inflammation and DNA damage in CRC. However, the experimental validations have been hindered by cost, resources, and cumbersome technical expertise required for metabolomic investigations. In this study, we performed a meta-analysis of CRC microbiota data from diverse geographical regions using multiple methods to achieve reproducible results. We used a computational approach to predict the metabolomic profiles using existing CRC metagenomic datasets. We identified a reliable set of CRC-specific biomarkers from this analysis, including microbial and metabolite markers. In addition, we revealed significant microbe-metabolite associations through correlation analysis and microbial gene families associated with dysregulated metabolic pathways in CRC, which are essential in understanding the vastly sporadic nature of CRC development and progression.
Keywords: biomarkers; colorectal cancer; gut dysbiosis; meta-analysis; microbial metabolites; microbiome.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
-
- Mokarram P, Albokashy M, Zarghooni M, Moosavi MA, Sepehri Z, Chen QM, Hudecki A, Sargazi A, Alizadeh J, Moghadam AR, Hashemi M, Movassagh H, Klonisch T, Owji AA, Łos MJ, Ghavami S. 2017. New frontiers in the treatment of colorectal cancer: autophagy and the unfolded protein response as promising targets. Autophagy 13:781–819. doi: 10.1080/15548627.2017.1290751. - DOI - PMC - PubMed
-
- Wu XC, Chen VW, Steele B, Ruiz B, Fulton J, Liu L, Carozza SE, Greenlee R. 2001. Subsite-specific incidence rate and stage of disease in colorectal cancer by race, gender, and age group in the United States, 1992–1997. Cancer 92:2547–2554. doi: 10.1002/1097-0142(20011115)92:10<2547::AID-CNCR1606>3.0.CO;2-K. - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

