A Method for Variant Agnostic Detection of SARS-CoV-2, Rapid Monitoring of Circulating Variants, and Early Detection of Emergent Variants Such as Omicron
- PMID: 35766514
- PMCID: PMC9297815
- DOI: 10.1128/jcm.00342-22
A Method for Variant Agnostic Detection of SARS-CoV-2, Rapid Monitoring of Circulating Variants, and Early Detection of Emergent Variants Such as Omicron
Abstract
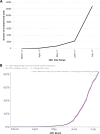
The rapid emergence of SARS-CoV-2 variants raised public health questions concerning the capability of diagnostic tests to detect new strains, the efficacy of vaccines, and how to map the geographical distribution of variants to understand transmission patterns and loads on healthcare resources. Next-generation sequencing (NGS) is the primary method for detecting and tracing new variants, but it is expensive, and it can take weeks before sequence data are available in public repositories. This article describes a customizable reverse transcription PCR (RT-PCR)-based genotyping approach which is significantly less expensive, accelerates reporting, and can be implemented in any lab that performs RT-PCR. Specific single-nucleotide polymorphisms (SNPs) and indels were identified which had high positive-percent agreement (PPA) and negative-percent agreement (NPA) compared to NGS for the major genotypes that circulated through September 11, 2021. Using a 48-marker panel, testing on 1,031 retrospective SARS-CoV-2 positive samples yielded a PPA and NPA ranging from 96.3 to 100% and 99.2 to 100%, respectively, for the top 10 most prevalent World Health Organization (WHO) lineages during that time. The effect of reducing the quantity of panel markers was explored, and a 16-marker panel was determined to be nearly as effective as the 48-marker panel at lineage assignment. Responding to the emergence of Omicron, a genotyping panel was developed which distinguishes Delta and Omicron using four highly specific SNPs. The results demonstrate the utility of the condensed panel to rapidly track the growing prevalence of Omicron across the US in December 2021 and January 2022.
Keywords: COVID-19; Delta; Omicron; RT-PCR; SARS-CoV-2; genotyping; in vitro diagnostics; mutations; next-generation sequencing; variants.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

References
-
- Wölfel R, Corman VM, Guggemos W, Seilmaier M, Zange S, Müller MA, Niemeyer D, Jones TC, Vollmar P, Rothe C, Hoelscher M, Bleicker T, Brünink S, Schneider J, Ehmann R, Zwirglmaier K, Drosten C, Wendtne C. 2020. Virological assessment of hospitalized patients with COVID-2019. Nature 581:465–469. 10.1038/s41586-020-2196-x. - DOI - PubMed
-
- Korber B, Fischer WM, Gnanakaran S, Yoon H, Theiler J, Abfalterer W, Hengartner N, Giorgi EE, Bhattacharya T, Foley B, Hastie KM, Parker MD, Partridge DG, Evans CM, Freeman TM, de Silva TI, McDanal C, Perez LG, Tang H, Moon-Walker A, Whelan SP, LaBranche CC, Saphire EO, Montefiori DC., Sheffield COVID-19 Genomics Group. 2020. Tracking changes in SARS-CoV-2 spike: evidence that D614G increases infectivity of the COVID-19 virus. Cell 182:812–827.e19. 10.1016/j.cell.2020.06.043. - DOI - PMC - PubMed
-
- Hodcroft EB, Domman DB, Snyder DJ, Oguntuyo KY, Van Diest M, Densmore KH, Schwalm KC, Femling J, Carroll JL, Scott RS, Whyte MM, Edwards MW, Hull NC, Kevil CG, Vanchiere JA, Lee B, Dinwiddie DL, Cooper VS, Kamil JP. 2021. Emergence in late 2020 of multiple lineages of SARS-CoV-2 Spike protein variants affecting amino acid position 677. medRxiv 10.1101/2021.02.12.21251658. - DOI
Publication types
MeSH terms
Supplementary concepts
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

