Radiomics in Abdominopelvic Solid-Organ Oncologic Imaging: Current Status
- PMID: 35766531
- PMCID: PMC10616929
- DOI: 10.2214/AJR.22.27695
Radiomics in Abdominopelvic Solid-Organ Oncologic Imaging: Current Status
Abstract
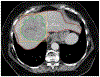
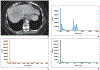
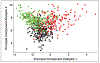
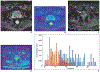
Radiomics is the process of extraction of high-throughput quantitative imaging features from medical images. These features represent noninvasive quantitative biomarkers that go beyond the traditional imaging features visible to the human eye. This article first reviews the steps of the radiomics pipeline, including image acquisition, ROI selection and image segmentation, image preprocessing, feature extraction, feature selection, and model development and application. Current evidence for the application of radiomics in abdominopelvic solid-organ cancers is then reviewed. Applications including diagnosis, subtype determination, treatment response assessment, and outcome prediction are explored within the context of hepatobiliary and pancreatic cancer, renal cell carcinoma, prostate cancer, gynecologic cancer, and adrenal masses. This literature review focuses on the strongest available evidence, including systematic reviews, meta-analyses, and large multicenter studies. Limitations of the available literature are highlighted, including marked heterogeneity in radiomics methodology, frequent use of small sample sizes with high risk of overfitting, and lack of prospective design, external validation, and standardized radiomics workflow. Thus, although studies have laid a foundation that supports continued investigation into radiomics models, stronger evidence is needed before clinical adoption.
Keywords: abdomen; features; oncology; radiomics; texture.
Figures


References
-
- Pinto Dos Santos D. Radiomics in endometrial cancer and beyond—a perspective from the editors of the EJR. Eur J Radiol 2022; 150:110266. - PubMed
-
- Croxen J, Kaganoff S. Pulse check: an analysis of the FDA’s list of AI- and ML-enabled devices. rockhealth.com/insights/pulse-check-an-analysis-of-the-fdas-list-of-ai-a.... Published October 8, 2021. Accessed May 29, 2022
-
- Lambin P, Leijenaar RTH, Deist TM, et al. Radiomics: the bridge between medical imaging and personalized medicine. Nat Rev Clin Oncol 2017; 14:749–762 - PubMed
-
- Bates SE. It’s all about the test: the complexity of companion diagnostic co-development in personalized medicine. Clin Cancer Res 2014; 20:1418. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

