Quantification of Protein Glycosylation Using Nanopores
- PMID: 35766994
- PMCID: PMC9284675
- DOI: 10.1021/acs.nanolett.2c01338
Quantification of Protein Glycosylation Using Nanopores
Abstract
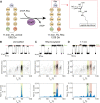
Although nanopores can be used for single-molecule sequencing of nucleic acids using low-cost portable devices, the characterization of proteins and their modifications has yet to be established. Here, we show that hydrophilic or glycosylated peptides translocate too quickly across FraC nanopores to be recognized. However, high ionic strengths (i.e., 3 M LiCl) and low pH (i.e., pH 3) together with using a nanopore with a phenylalanine at its constriction allows the recognition of hydrophilic peptides, and to distinguish between mono- and diglycosylated peptides. Using these conditions, we devise a nanopore method to detect, characterize, and quantify post-translational modifications in generic proteins, which is one of the pressing challenges in proteomic analysis.
Keywords: nanopore spectrometry; protein glycosylation; proteomics; rhamnosylation; single molecule.
Conflict of interest statement
The authors declare the following competing financial interest(s): G.M. is a founder, director, and shareholder of Portal Biotech Limited, a company engaged in the development of nanopore technologies. This work was not supported by Portal Biotech Limited.
Figures


References
-
- Roth Z.; Yehezkel G.; Khalaila I. Identification and Quantification of Protein Glycosylation. Int. J. Carbohydr. Chem. 2012, 2012, 1–10. 10.1155/2012/640923. - DOI
-
- West M. B.; Partyka K.; Feasley C. L.; Maupin K. A.; Goppallawa I.; West C. M.; Haab B. B.; Hanigan M. H. Detection of Distinct Glycosylation Patterns on Human γ-Glutamyl Transpeptidase 1 Using Antibody-Lectin Sandwich Array (ALSA) Technology. BMC Biotechnol. 2014, 14 (1), 101.10.1186/s12896-014-0101-0. - DOI - PMC - PubMed
-
- Zaro B. W.; Yang Y.-Y.; Hang H. C.; Pratt M. R. Chemical Reporters for Fluorescent Detection and Identification of O-GlcNAc-Modified Proteins Reveal Glycosylation of the Ubiquitin Ligase NEDD4–1. Proc. Natl. Acad. Sci. U. S. A. 2011, 108 (20), 8146–8151. 10.1073/pnas.1102458108. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

